Fig. 4
- ID
- ZDB-FIG-241016-58
- Publication
- Lascaux et al., 2024 - TEX264 drives selective autophagy of DNA lesions to promote DNA repair and cell survival
- Other Figures
- All Figure Page
- Back to All Figure Page
|
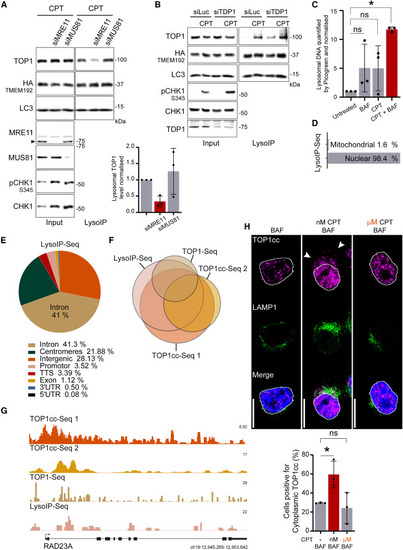
TOP1cc (TOP1 and its bound DNA fragment) are processed by lysosomes (A) LysoIP performed after 5 h of 50 nM CPT, and quantification graph (n = 3). Error bar, SD. (B) LysoIP performed after 5 h of 50 nM CPT (n = 3). (C) Quantification of DNA by Picogreen assay to detect DNA purified by LysoIP after 6 h of treatment (n = 3). Two-way ANOVA. Error bar, SD. Around 450 ng of DNA fragments of less than 1 kb were purified from 20 million cells by LysoIP after treatment with 50 nM CPT and 50 nM BAF. (D) Distribution of the LysoIP-purified DNA, mapped reads sequenced by NGS. (E) Genomic distribution of the LysoIP-purified mapped reads sequenced by NGS. (F) Proportional Eulerr representation of the overlap among the different ChIP-seq of TOP1cc, TOP1cc-seq 1 (MCF7—GEO: GSE135808),87 TOP1cc-seq 2 (LNCAP—GEO: GSE135808),87 and TOP1-seq (HCT116—GEO: GSE57628).88 All conditions were treated with CPT. (G) Genomic browser alignment on genome hg38 of the different TOP1cc ChIP-seq and LysoIP-seq at RAD23A gene loci as sequence tags per million (TPM). (H) Immunofluorescence after 4 h of 50 nM CPT (nM) or 1 μM CPT (μM) and quantification (n = 3). Scale bar, 10 μm. Two-way ANOVA. ∗p < 0.05; ns, not significant. See also Figure S4. |
Reprinted from Cell, 187(20), Lascaux, P., Hoslett, G., Tribble, S., Trugenberger, C., Antičević, I., Otten, C., Torrecilla, I., Koukouravas, S., Zhao, Y., Yang, H., Aljarbou, F., Ruggiano, A., Song, W., Peron, C., Deangeli, G., Domingo, E., Bancroft, J., Carrique, L., Johnson, E., Vendrell, I., Fischer, R., Ng, A.W.T., Ngeow, J., D'Angiolella, V., Raimundo, N., Maughan, T., Popović, M., Milošević, I., Ramadan, K., TEX264 drives selective autophagy of DNA lesions to promote DNA repair and cell survival, 5698-5718.e26, Copyright (2024) with permission from Elsevier. Full text @ Cell