- Title
-
Reactivation of an embryonic cardiac neural crest transcriptional profile during zebrafish heart regeneration
- Authors
- Dhillon-Richardson, R.M., Haugan, A.K., Lyons, L.W., McKenna, J.K., Bronner, M.E., Martik, M.L.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
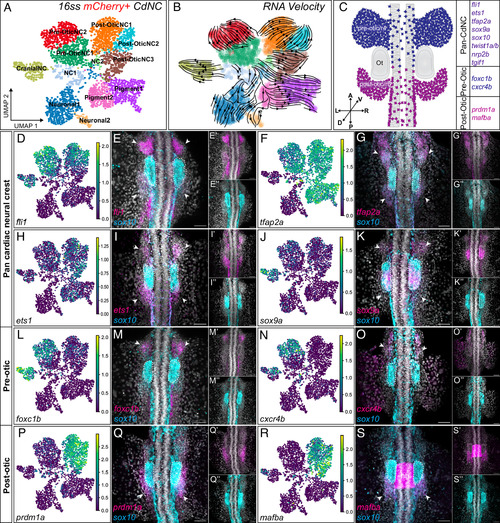
Single-cell analysis reveals two transcriptionally distinct populations of CdNC. (A) Labeled leiden clustering UMAP of scRNA sequencing of FAC sorted mCherry+ CdNC cells. (B) RNA velocity embedded on UMAP. (C) Illustration of a dorsal view, 16ss zebrafish embryo with pre- and postotic CdNC domains labeled and validated genes listed that are restricted to either domain or shared among both domains. (D–S″) UMAPs for individual genes and corresponding 20× confocal images of HCR expression patterns of dorsal view, 16ss zebrafish embryos, showing overlap in expression and individual channel insets of sox10 in cyan and the following genes of interest in magenta: (D–E″) fli1 (n = 11), (F–G″) tfap2a (n = 8), (H–I″) ets1 (n = 8), (J–K″) sox9a (n = 6), (L–M″) foxc1b (n = 3), (N–O″) cxcr4b (n = 8), (P–Q″) prdm1a (n = 6), and (R–S″) mafba (n = 15). The white arrows indicate expression in preotic and/or postotic CdNC streams, and the white dotted circles outline the otic vesicle. Acronyms: NC, neural crest; r4, rhombomere 4; r6, rhombomere 6; Ot, otic vesicle. Position: A, anterior; P, posterior; D, dorsal; V, ventral; L, left; R, right; hb, hindbrain. (Scale bar, 50 μm.) EXPRESSION / LABELING:
|
|
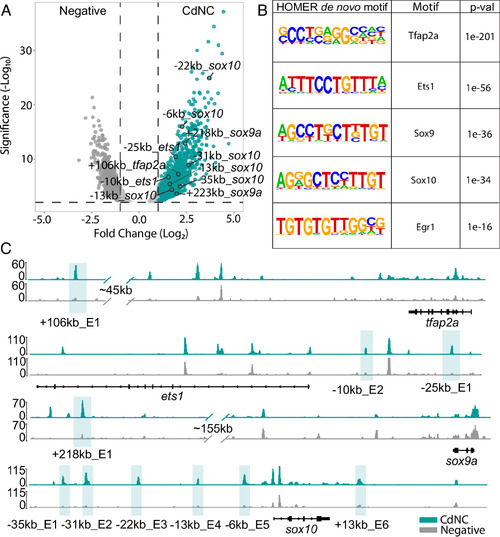
ATAC-seq and differential chromatin accessibility and transcription factor motif analysis uncovers putative enhancers and transcriptional regulators unique to the CdNC. (A) Volcano plot of merged peaks of sorted bulkATAC-seq shows differentially accessible regions in CdNC (mCherry+) and non-NC/negative cells (mCherry-). (B) Results of HOMER de novo motif analysis showing motif, transcription factor, and associated P-value. (C) IGV genome browser tracks of the ets1, tfap2a, sox9a, and sox10 loci. Highlighted peaks represent putative enhancer regions called by Diffbind (turquoise track = CdNC, gray track = negative). |
|
Loss of egr1 leads to a downregulation of CdNC genes and suggests an important role as core CdNC subcircuit transcription factor. HCR staining for ets1 and sox9a in 16ss zebrafish embryos injected with negative control gRNA (n = 11) (A and A”) and egr1 gRNA (n = 9) (B and B”). HCR staining for tfap2a and sox10 in 16ss zebrafish embryos injected with negative control gRNA (n = 9) (C and C″) and egr1 gRNA (n = 14) (D and D”). (E–H) Quantification of CTCF in negative control and egr1 gRNA injected embryos (unpaired Student’s t test P < 0.05). (I) Migratory CdNC subcircuit displaying proposed interactions between shown transcription factors and downstream enhancer regions. (Scale bar, 50 μm.) EXPRESSION / LABELING:
PHENOTYPE:
|
|
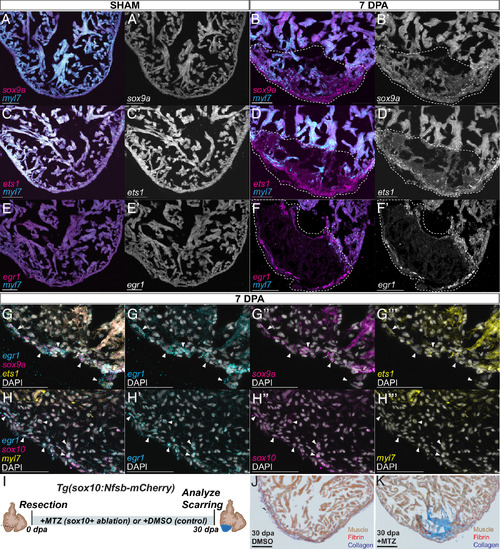
A developmental CdNC subcircuit is expressed during adult cardiac regeneration. HCR staining of sham and injured 7dpa ABWT hearts, respectively, for myl7 and (A and B) sox9a (n = 12/12), (C and D) ets1 (n = 11/11), and (E and F) egr1 (n = 15/15). (A′–F′) Grayscale of respective sox9a, ets1, and egr1 stainings. (Scale bar, 100 μm.) (G) HCR costaining on ABWT 7dpa hearts for sox9a, egr1, ets1, and DAPI (n = 8/8). Arrows indicate some of the cells positive for all three probes. Single channel images, with DAPI, of, sox9a, egr1, and ets1 are shown respectively in (G′ and G″′). (Scale bar, 50 μm.) (H) HCR costaining on ABWT 7dpa hearts for sox10, egr1, myl7, and DAPI (n = 10/11). Arrows indicate a subset of the cells double-positive for sox10 and egr1 probes. Single channel images, with DAPI, of egr1, sox10, and myl7 are shown respectively in (H′ and H″). (I) Diagram of the sox10+ ablation scheme. Tg(sox10:Nfsb-mCherry) injured fish are maintained in MTZ for the duration of the regeneration time course and hearts are harvested at 30 dpa. (J) Injured hearts maintained in DMSO to 30 dpa retain little scarring (n = 3/3), whereas (K) injured hearts maintained in MTZ, ablating sox10+ cells, maintain a scar to 30 dpa (n = 5/5). (Scale bar, 100 μm.) |