- Title
-
Protease-mediated activation of Par2 elicits calcium waves during zebrafish egg activation and blastomere cleavage
- Authors
- Ma, J., Carney, T.J.
- Source
- Full text @ PLoS Biol.
|
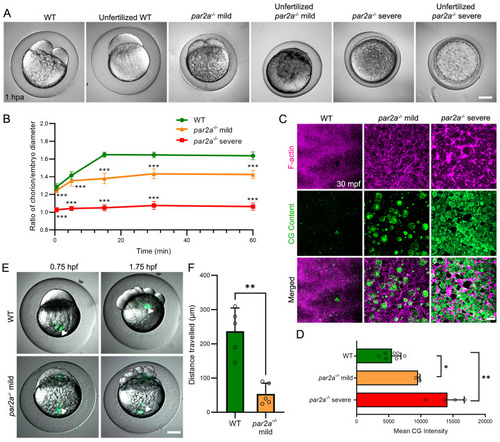
Serine protease inhibitors cause egg activation defects and reduced calcium waves. PHENOTYPE:
|
|
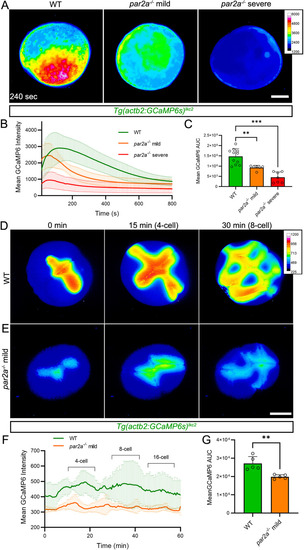
Maternal PHENOTYPE:
|
|
PHENOTYPE:
|
|
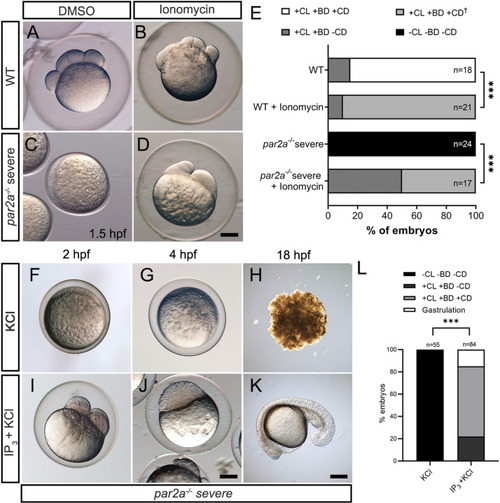
Increasing intracellular Ca2+ either by Ionomycin or IP3 rescues egg activation defects in PHENOTYPE:
|
|
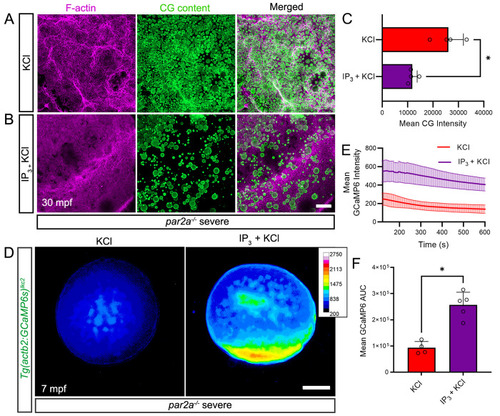
IP3 rescues egg activation defects by increasing intracellular calcium spatially. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|