- Title
-
Amylovis-201 is a new dual-target ligand, acting as an anti-amyloidogenic compound and a potent agonist of the σ1 chaperone protein
- Authors
- García-Pupo, L., Crouzier, L., Bencomo-Martínez, A., Meunier, J., Morilleau, A., Delprat, B., Carrazana, M.S., Menéndez Soto Del Valle, R., Maurice, T., Rodríguez-Tanty, C.
- Source
- Full text @ Acta Pharm Sin B
|
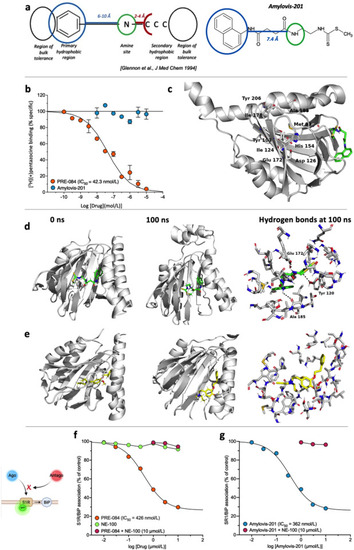
Amylovis-201 is a σ1 receptor interacting drug. (a) Pharmacophore structure proposed by Glennon et al.14 and application to the developed formula of Amylovis-201. Note that the secondary hydrophobic region does not fit with the hydrophilic dithiocarbamate part. (b) Inhibition of (+)-[3H]pentazocine specific binding to Jurkat human leukemic T cells by PRE-084 and Amylovis-201. (c) Amino acids identified in the ligand binding pocket (LBP) of the σ1 protein and which are critical for ligand-protein interactions. In the 3D model of the protein, the LBP is profoundly occluded within the protein and unreachable for Amylovis-201. Molecular dynamics were performed between 0 and 100 ns with (d) Amylovis-201 and (e) NE-100. H-bonds are shown as dashed lines at 100 ns (f) PRE-084 dissociated BiP from σ1 protein in GFP-tagged σ1 protein-overexpressing CHO cells with an IC50 value of 426 nmol/L. NE-100 failed to do so at concentrations up to 10 μmol/L, but, at 10 μmol/L, prevented the dissociating effect of PRE-084. (g) Amylovis-201 dissociated BiP from σ1 protein with an IC50 of 362 nmol/L. Its effect was blocked by NE-100. Experiments were performed in duplicates. |
|
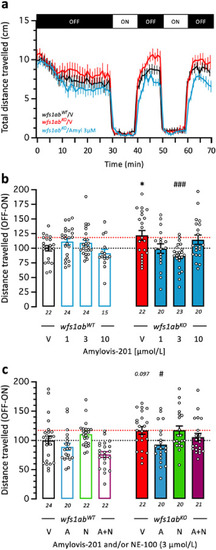
Amylovis-201 attenuated the hyperlocomotor response of 5 dpfwfs1abKO mutant zebrafish larvae in the VMR test through σ1 agonism: (a) VMR activity profiles of V-treated wfs1abWTand wfs1abKO mutant lines and wfs1abKO zebrafish treated with Amylovis-201 at 3 μmol/L; (b) dose–response effect of Amylovis-201 on the zebrafish mobility; and (c) effect of NE-100. The fish mobility was measured for 70 min, with 30 min of training in the dark (OFF), then 2 cycles of light/dark (ON/OFF) of 10 min each. In (b, c), the distance traveled by wfs1abWT and wfs1abKO larvae during the light/dark cycle was averaged as differences between the OFF and ON phases. Data show mean ± SEM of the number of animals indicated below the columns. Two-way ANOVAs: F(1,162) = 0.4218, P = 0.5170 for the genotype, F(3,162) = 1.316, P = 0.2710 for the treatment, F(3,162) = 5.561, P = 0.0012 for the interaction in (b); F(1,163) = 9.608, P = 0.0023 for the genotype, F(3,63) = 6.781, P = 0.0002 for the treatment, F(3,163) = 1.616, P = 0.1876 for the interaction in (c). ∗P < 0.05 vs. V-treated wfs1abWT zebrafish; #P < 0.05, ###P < 0.001 vs. V-treated wfs1abKO zebrafish; Dunnet's test. |
|
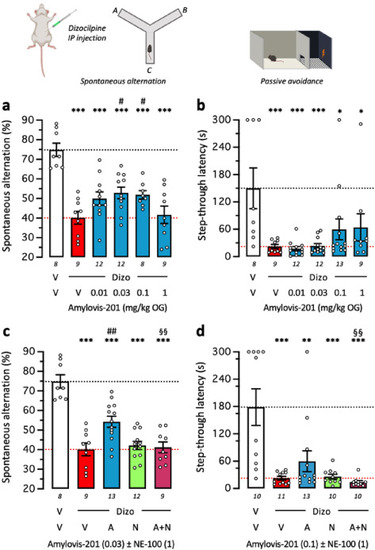
Amylovis-201 attenuated Dizocilpine-induced learning impairment in mice through σ1 agonism: dose-response effects in the (a) spontaneous alternation and (b) step-through passive avoidance. (c,d) Blockade of Amylovis-201 effect by NE-100 in each test. Mice received Amylovis-201 (0.01–1 mg/kg PO) 10 min before Dizocilpine (0.15 mg/kg IP), 20 min before the Y-maze session in (a,c) or passive avoidance training in (b,d). In (c,d), NE-100 was administered at 1 mg/kg IP simultaneously with the lowest active dose of Amylovis-201, i.e., 0.03 mg/kg PO in the Y-maze in (d) and 0.1 mg/kg in the passive avoidance test in (e). Abbreviations: V, vehicle solution (physiological saline); Dizo, Dizocilpine; A, Amylovis-201; N, NE-100. Data show mean ± SEM of the number of animals indicated within or below the columns. ANOVAs: F(5,50) = 12.19, P < 0.0001 in (a); F(4,46) = 22.76, P < 0.0001 in (c). Kruskal-Wallis ANOVAs: H = 19.29, P = 0.0017 in (b); H = 23.86, P < 0.0001 in (d). ∗ P < 0.05, ∗∗ P < 0.01, ∗∗∗ P < 0.001 vs. (V+V)-treated mice; # P < 0.05, ## P < 0.01 vs. (Dizo+V)-treated mice; oo P < 0.01 vs. (Dizo+A)-treated mice; Dunnett's test in (a,c), Dunn's test in (b,d). |
|
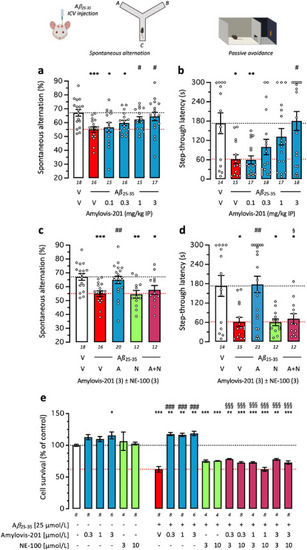
Amylovis-201 attenuated Aβ25–35-induced learning impairment in mice and Aβ25–35-induced cytotoxicity in SH-SY5Y cells through σ1 agonism: dose–response effects in the (a) spontaneous alternation in the Y-maze and (b) step-through passive avoidance. (c, d) Blockade of Amylovis-201 effect by NE-100 in each test. Mice received oligomerized Aβ25–35 peptide (9 nmol, icv) or vehicle solution and then Amylovis-201 (0.3–3 mg/kg ip) 7 days before behavioral analyses. The Y-maze session was performed on Day 7 (a, c) and passive avoidance training on Day 8 and retention on Day 9 (b, d). In (c, d), NE-100 was administered at 3 mg/kg ip simultaneously with Amylovis-201 at 3 mg/kg IP. (e) SH-SY5Y neuroblastoma cells (10,000 cells/well) were exposed to Aβ25–35 (25 μmol/L) and Amylovis-201 (0.3–3 μmol/L) and/or NE-100 (3, 10 μmol/L). Cell survival was monitored after 24 h using the MTT assay. Abbreviations: V, vehicle solution (physiological saline); A, Amylovis-201; N, NE-100. Data show mean ± SEM of the number of determination indicated within or below the columns. ANOVAs: F(5,90) = 3.336, P = 0.0082 in (a); F(4,72) = 5.109, P = 0.0011 in (c); F(11,80) = 49.51, P < 0.0001 in (e). Kruskal–Wallis ANOVAs: H = 13.5, P = 0.0019 in (b); H = 14.18, P = 0.007 in (d). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. (V + V)-treated mice or untreated cells; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. (Aβ25–35+V)-treated mice or cells; §P < 0.05, §§§P < 0.001 vs. (Aβ25–35 +Amylovis-201)-treated mice or cells; Dunnett's test in (a, c, e), Dunn's test in (b, d). |
|
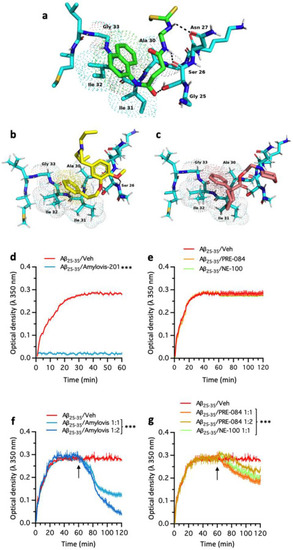
Amylovis-201 anti-aggregating properties in vitro. Molecular docking with Aβ25–35 peptide. Molecular rigid docking models of the complexes (stick representations): (a) Amylovis-201-Aβ25–35 peptide; (b) NE-100-Aβ25–35 peptide; (c) PRE-084-Aβ25–35 peptide. The Aβ25–35 amyloid peptide code used 1QXC (PDB). (d) Light scattering of Aβ25–35 (25 μmol/L in 40 mmol/L HEPES, pH 7.4) spectrophotometrically monitored at 43 °C in absence or presence of Amylovis-201. Numbers in parentheses indicate molar ratios of proteins. (e) Light scattering of Aβ25–35 in absence or presence of PRE-084 or NE-100. (f) Light scattering of Aβ25–35 when Amylovis-201, PRE-084 or NE-100 were added after 1 h at a stoichiometry 1:1. (g) Same experiment with a stoichiometry1:2. Experiments were performed in triplicates. |