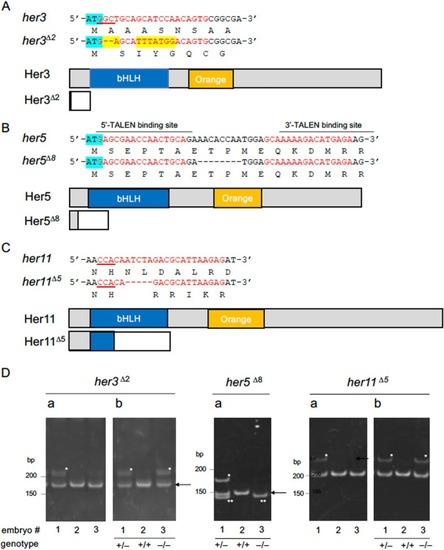
Structures of the mutant her genes established in the current study. (A–C) Mutations introduced into Notch‐independent her genes by genome editing. Small indels were introduced into the N‐termini (her3, her5) or bHLH (her11) domains using CRISPR/Cas9 (her3, her11) or TALEN (her5). The target base sequences (red) and corresponding amino acid sequences are shown above the gene schematics. PAM sequences are underlined, and start codons are marked with light blue. In her3, base substitution (yellow) and a small deletion were observed. Wild‐type and disrupted coding sequences due to frameshifts are shown with gray and white boxes, except for bHLH domains and Orange domains that are shown with blue and orange boxes, respectively. In the her3 mutant line, almost the entire coding region was missing due to a frameshift immediately after the start codon. In her5 mutants, an 8‐bp deletion resulted in a stop codon at position 29. In the her11 mutant, a 5‐bp deletion occurred in the N‐terminal portion of the bHLH domain. (D) Genotyping of her mutants by the HMA method (examples). Da. Genomic DNA from embryos (#1–#3) was PCR amplified using primer pairs flanking the target sites and analyzed by PAGE. Band shifts (asterisks) indicate heterozygotes, while no shift indicates WT or homozygotes. In her5 mutants, WT and homozygotes were discriminated by size reduction (double asterisks). For her3/her11 mutants, PCR products were re‐annealed with WT genomic DNA and analyzed by second PAGE (Db). Band shifts confirm homozygosity.
|

