- Title
-
The transcription factors Tfeb and Tfe3 are required for survival and embryonic development of pancreas and liver in zebrafish
- Authors
- Rissone, A., La Spina, M., Bresciani, E., Syed, Z.A., Combs, C.A., Kirby, M., Elkahloun, A., Chen, V., Sood, R., Burgess, S.M., Puertollano, R.
- Source
- Full text @ PLoS Genet.
|
Generation of zebrafish |
|
Proteomic and phenotypical analysis of 5 dpf WT and TKO embryos. EXPRESSION / LABELING:
|
|
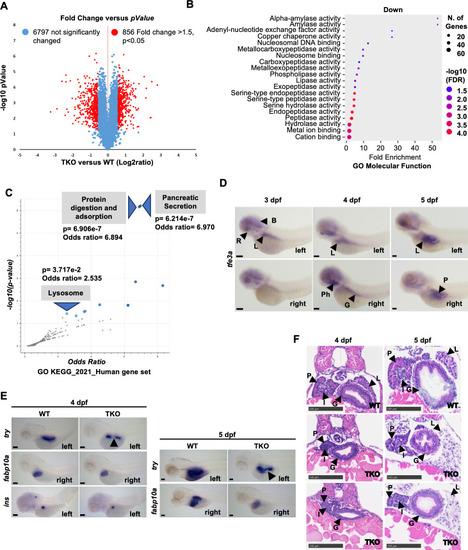
Pancreatic defects observed in TKO embryos and adult fish. |
|
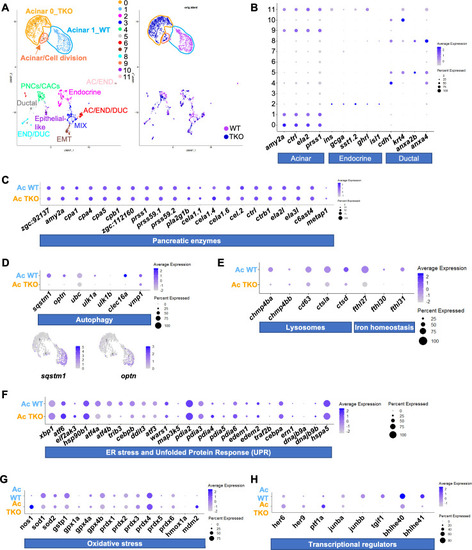
Single cell RNA-sequencing of 4 dpf WT and TKO GFP |
|
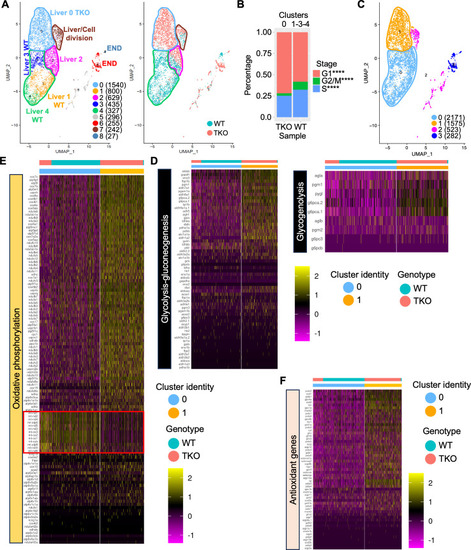
Liver and gut defects observed in TKO embryos. |
|
Single cell RNA-sequencing of 4 dpf WT and TKO dsRED |
|
TKO embryos show higher sensitivity to different kinds of cellular stress. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|