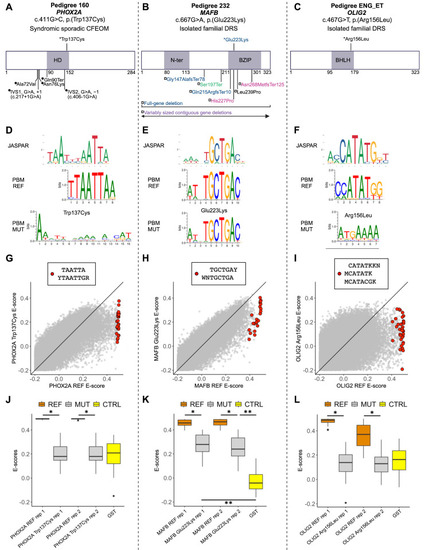
The oCCDD proband-derived candidate variants in transcription factors disrupt DNA binding. (A–C) Two-dimensional structural mapping of human variants. (A) Variants in PHOX2A associated with CFEOM. (B) Variants in MAFB associated with DRS and/or co-occurring phenotypes. (C) Variant in OLIG2 associated with DRS. References for Figure 7 are provided in Supplementary Methods and Results. Asterisks represent variants above schematics that were identified and reported in our sequenced human oCCDD cohort1 and are functionally tested for the first time in this work: PHOX2A p.(Trp137Cys), MAFB p.(Glu223Lys), and OLIG2 p.(Arg156Leu). Variants below schematics are previously reported as follows: (A) Filled circles represent previously reported PHOX2A variants associated with CFEOM: IVS1, G>A, +1 (c.217+1G>A)2; IVS2, G>A, −1(c.406-1G>A)2,3; p.(Ala72Val)2,3; p.(Asn76Lys)4; p.(Gln90Ter).5 (B) -Previously reported MAFB variants associated with DRS and colored as follows: blue denotes isolated DRS6; magenta denotes DRS ± hearing impairment ± intellectual disability6,7; black denotes DRS + focal segmental glomerulosclerosis ± hearing impairment8; green denotes focal segmental glomerulosclerosis ± DRS9; purple denotes contiguous gene deletions with variable neurodevelopmental anomalies ± DRS.10 Variants are mapped using the following transcripts: ENST00000298231.5 (PHOX2A), ENST00000373313.3 (MAFB), and ENST00000382357.4 (OLIG2). (D–F) Transcription factor binding site motif logos for PHOX2A (D), MAFB (E), and OLIG2 (F). Top: logos from the JASPAR database of transcription factor binding profiles derived from high-throughput sequencing SELEX (HT-SELEX) experiments for either the wild-type human protein (PHOX2A and OLIG2) or orthologous mouse protein (Mafb). Middle and bottom: Protein binding microarray (PBM) experiment motifs derived from universal protein binding microarrays for the reference DNA binding domain (PBM REF, middle) and for the mutant DNA binding domain (PBM MUT, bottom). (G–I) For 8-mers resembling the wild-type motif, E-score comparison between reference and mutant DNA binding domain for PHOX2A (G), MAFB (H), and OLIG2 (I). Red dots correspond to 8-mer sequences that contain the labeled International Union of Pure and Applied Chemistry code-based k-mers, which are selected to closely resemble each motif logo. Y, C or T nucleotides; R, A or G nucleotides; W, A or T nucleotides; M, A or C nucleotides; K, G or T nucleotides; N, any base. (J–L) The 8-mer E-score comparison for the k-mers labeled in G–I across the replicate protein binding microarrays. (J) Reference versus p.(Trp137Cys) PHOX2A variant. PHOX2A p.(Trp137Cys) led to a significant drop in E-scores for 8-mers resembling the wild-type motif (P < 10−9; one-sided Mann-Whitney U test) to a level indistinguishable from E-scores of the GST negative control (P > 0.9; two-sided Mann-Whitney U test). (K) Reference versus p.(Glu223Lys) MAFB variant. The E-scores for the 8-mers recognized by the wild-type MAFB showed significantly lower values for the mutant DNA binding domain (P < 10−7; one-sided Mann-Whitney U test), but the E-score distribution for the mutant was still higher than that of the GST-negative control (P < 10−6; two-sided Mann-Whitney U test), suggesting partial loss of binding. (L) Reference versus p.(Arg156Leu) OLIG2 variant. OLIG2 p.(Arg156Leu) led to a significant reduction in mutant E-scores for 8-mers recognized by wild-type OLIG2 (P < 10−10; one-sided Mann-Whitney U test) to a level similar to that of the GST-tagged negative control (P > 0.1; two-sided Mann-Whitney U test). Orange, reference protein; gray, mutant/nonreference protein; yellow, GST-tagged negative control. * P < 1 × 10−7; one-sided Mann-Whitney U test. ** P < 1 × 10−6; two-sided Mann-Whitney U test. BHLH, basic helix-loop-helix domain; BZIP, basic leucine zipper domain; CFEOM, congenital fibrosis of the extraocular muscles; CTRL, GST-negative control protein; DRS, Duane retraction syndrome; GS, glutathione S-transferase tagged negative control protein; HD, homeodomain; MUT, mutant/nonreference; N-ter, N-terminal region; PBM, protein-binding microarray; REF, reference (nonmutant) sequence; rep, replicate.
|

