- Title
-
Zglp-1 is a novel essential transcriptional regulator for sex reversal in zebrafish
- Authors
- Wang, Y., Xu, G., Li, H., Gao, J., Du, X., Jiang, W., Ji, G., Liu, Z.
- Source
- Full text @ Mar Life Sci Technol
|
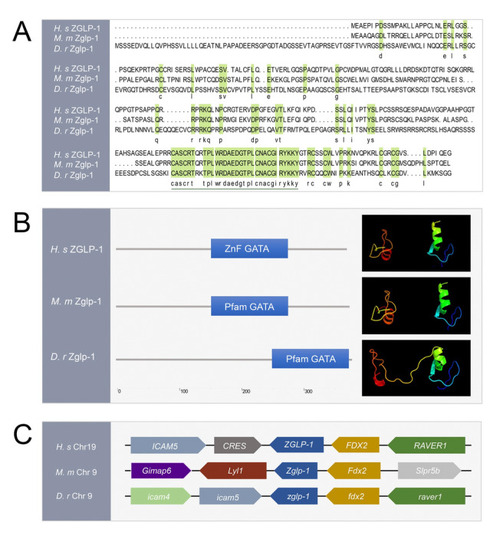
Zglp-1 zinc finger domains are well conserved in vertebrates. |
|
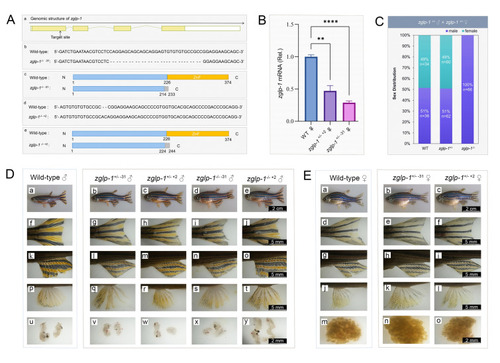
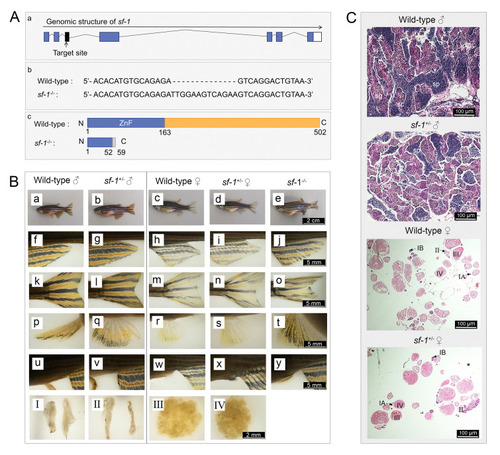
The lack of females observed in |
|
|
|
|
|
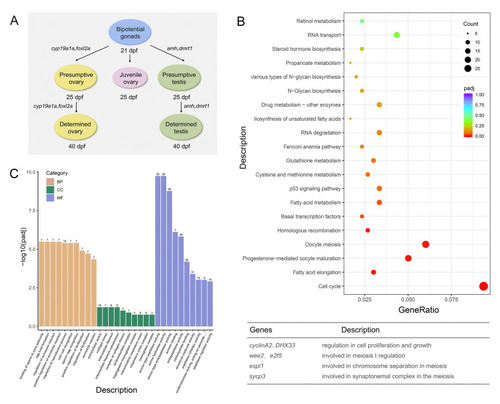
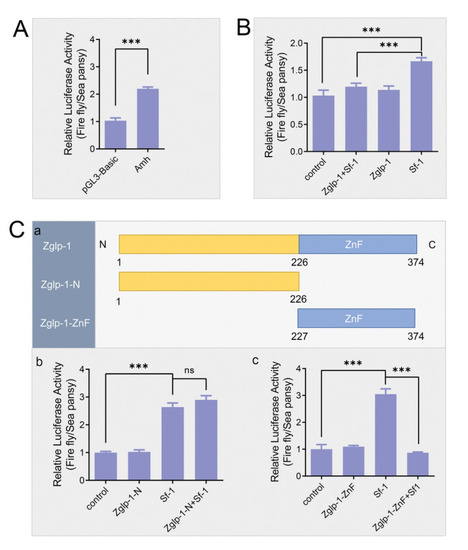
Zglp-1 inhibits the transcription of |
|
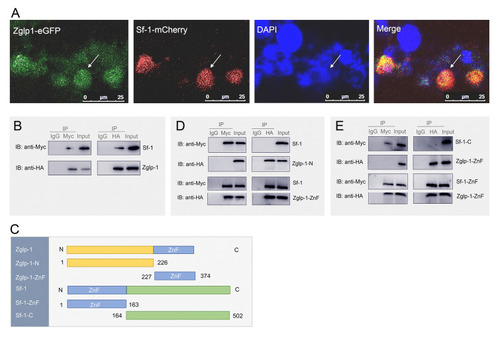
Interaction of Zglp-1 and Sf-1 occurs between their zinc finger domains. |
|
|
|
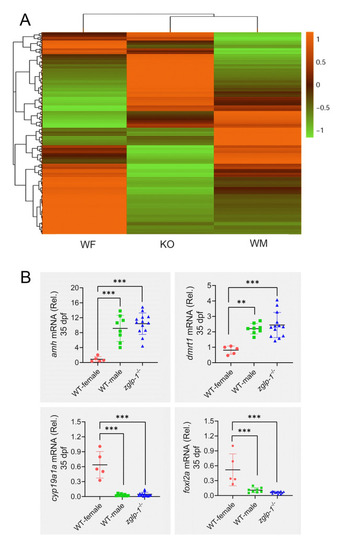
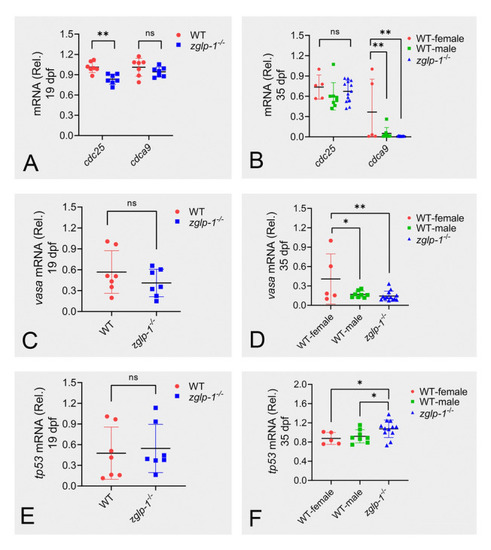
Zglp-1 regulates expression of genes involved in the proliferation and apoptosis of gonadal cells. |
|
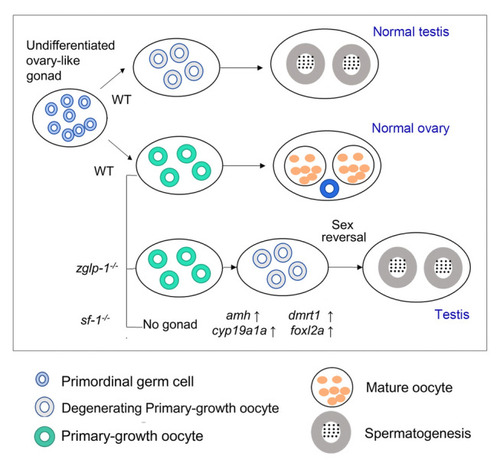
A model for the regulation of Zglp-1 in sex differentiation in zebrafish. The |