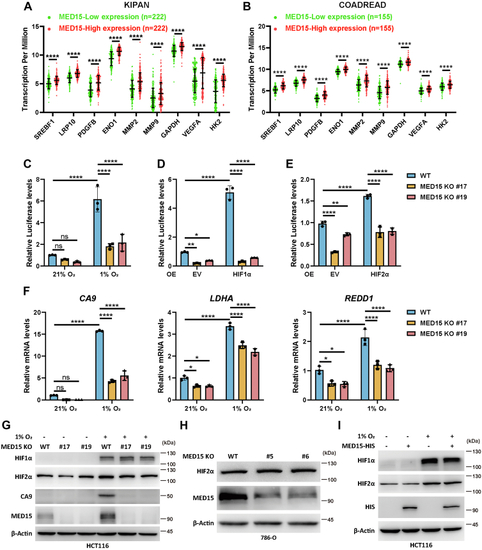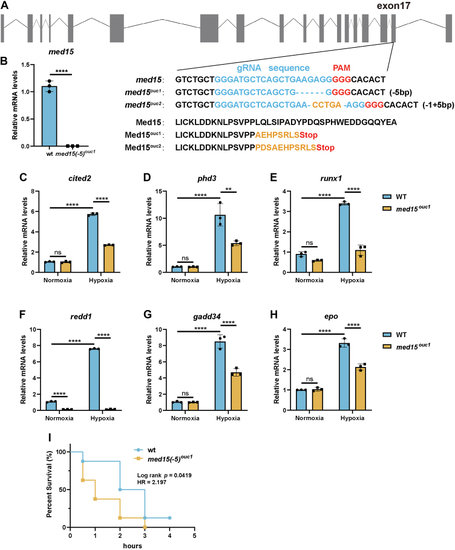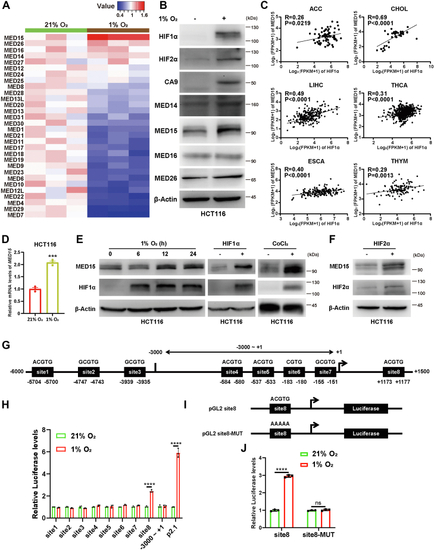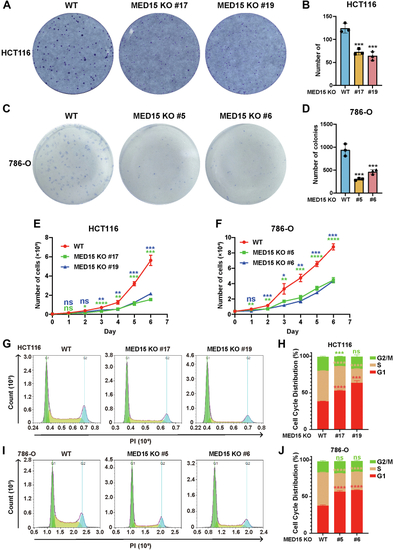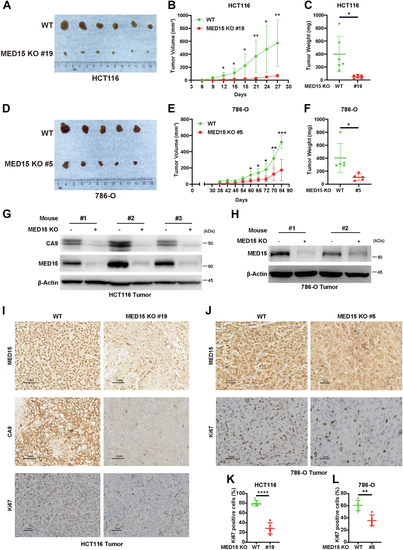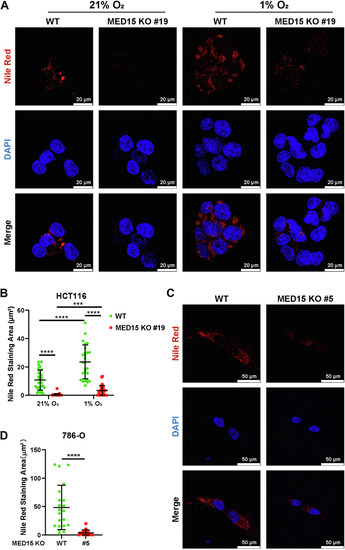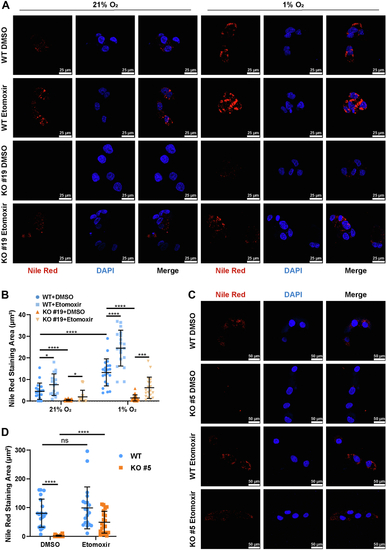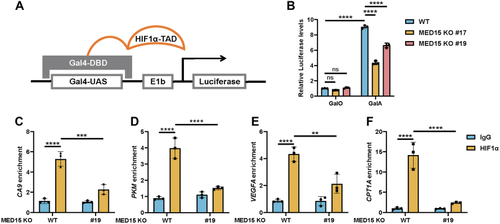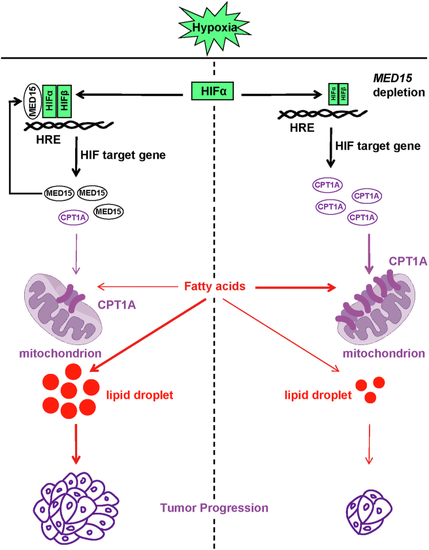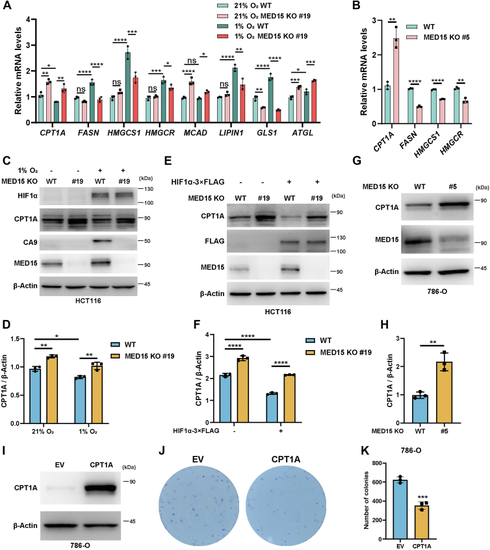
Knocking out MED15 activates the expression of CPT1A.A, RT-qPCR analysis of the mRNA levels of CPT1A, FASN, HMGCS1, HMGCR, MCAD, LIPIN1, GLS1, and ATGL in WT or MED15 KO (KO #19) HCT116 cells exposed to 21% O2 or 1% O2 for 72 h. Data are represented as means ± SD. n = 3 biologically independent extracts. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.0001, ∗∗∗∗p < 0.0001 by two-way ANOVA with Tukey's multiple comparisons test. B, RT-qPCR analysis of the the mRNA levels of CPT1A, FASN, HMGCS1, and HMGCR in WT or MED15 KO (KO #5) 786-O cells. Data are represented as means ± SD. n = 3 biologically independent extracts. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by Student's t test. C and D, Western blot of indicated proteins in WT or MED15 KO (KO #19) HCT116 cells exposed to 21% O2 or 1% O2 for 72 h (C). Relative intensities of CPT1A were quantified (D). Data are represented as means ± SD. n = 3 biologically independent extracts. ∗p < 0.05, ∗∗p < 0.01 by two-way ANOVA with Tukey's multiple comparisons test. E and F, Western blot of indicated proteins in WT or MED15 KO (KO #19) HCT116 cells transfected with EV (−) or HIF1α-3 × FLAG (+) (E). Relative intensities of CPT1A were quantified (F). Data are represented as means ± SD. n = 3 biologically independent extracts. ∗∗∗∗p < 0.0001 by two-way ANOVA with Tukey's multiple comparisons test. G and H, Western blot of indicated proteins in WT or MED15 KO (KO #5) 786-O cells (G). Relative intensities of CPT1A were quantified (H). Data are represented as means ± SD. n = 3 biologically independent extracts. ∗∗p < 0.01 by Student's t test. I, Western blot assay of CPT1A confirmed that CPT1A protein level significantly upregulated after stable overexpression of CPT1A. J and K, colony formation assay of 786-O with stable overexpression of EV or CPT1A. Cells were cultured for 9 days. Representative images from three independent experiments are shown in (J). Quantification of colony numbers is shown in (K). Data are represented as means ± SD. n = 3 biologically independent repeats. ∗∗∗p < 0.001 by Student's t test. ATGL, adipose triglyceride lipase; CPT1A, carnitine palmitoyltransferase 1A; FASN, fatty acid synthase; EV, empty vector; GLS1, glutaminase 1; HIF, hypoxia-inducible factor; HMGCS1, 3-hydroxy-3-methylglutaryl-CoA synthase 1; HMGCR, 3hydroxy-3methylglutaryl-coenzyme A reductase; MCAD, medium-chain specific acyl-CoA dehydrogenase; MED15, Mediator complex subunit 15; ns, not significant.
|