- Title
-
The inflammasome adaptor pycard is essential for immunity against Mycobacterium marinum infection in adult zebrafish
- Authors
- Uusi-Mäkelä, M., Harjula, S.E., Junno, M., Sillanpää, A., Nätkin, R., Niskanen, M.T., Saralahti, A.K., Nykter, M., Rämet, M.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
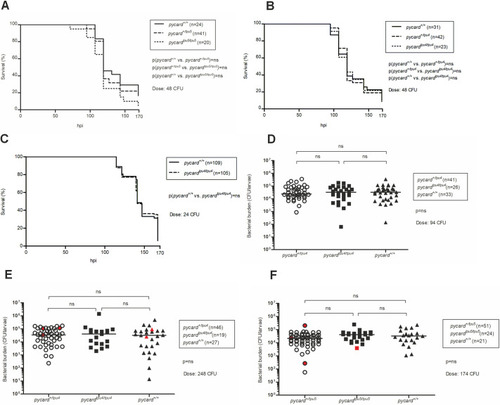
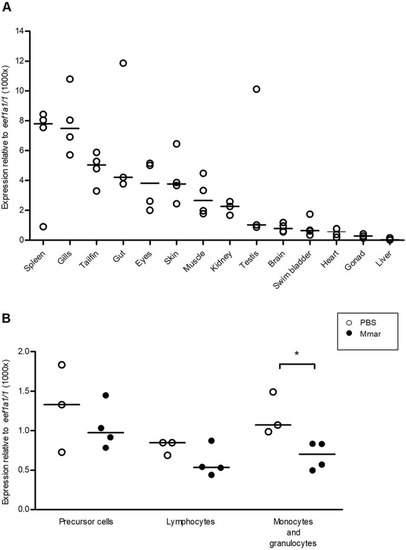
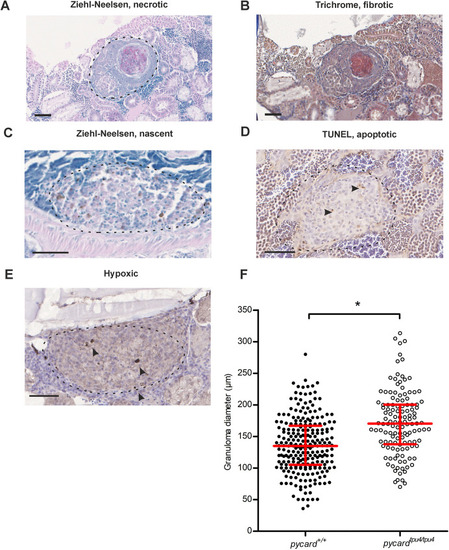
PHENOTYPE:
|
|
|
|
|
|
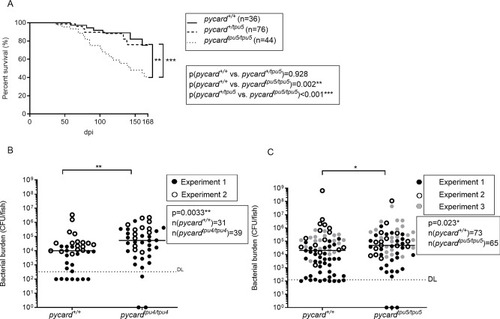
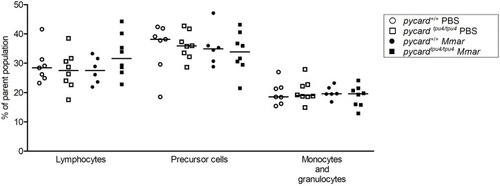
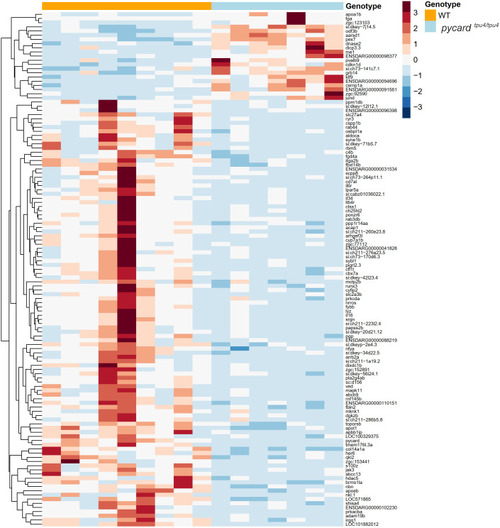
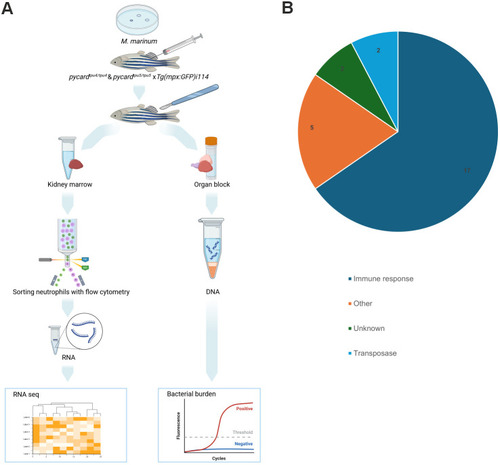
PHENOTYPE:
|
|
|
|
|
|
|