FIGURE
Fig. 1
- ID
- ZDB-FIG-250714-80
- Publication
- Tallan et al., 2025 - Highly quantitative measurement of differential protein-genome binding with PerCell chromatin sequencing
- Other Figures
- All Figure Page
- Back to All Figure Page
Fig. 1
|
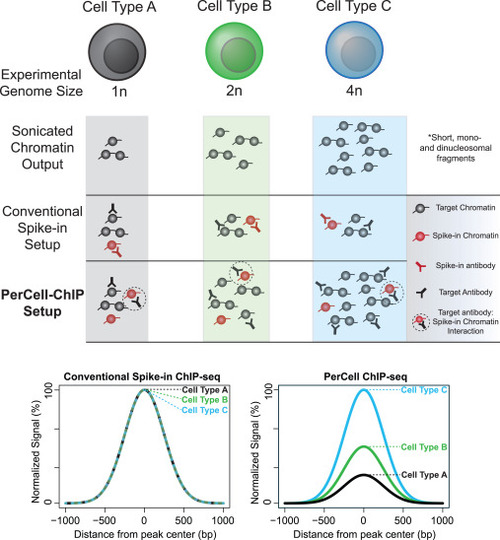
Rationale for PerCell-normalized semi-automated ChIP-seq analysis Shown are conceptual representations and comparisons of ChIP results after analyzing cells of varying ploidy (1n, 2n, and 4n) via conventional spike-in or PerCell normalization. The PerCell methodology specifically enables the detection of changes in relative signal across cellular backgrounds with different chromatin content and the use of a single antibody. |
Expression Data
Expression Detail
Antibody Labeling
Phenotype Data
Phenotype Detail
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep Methods