Fig. 3
- ID
- ZDB-FIG-250425-3
- Publication
- Yang et al., 2025 - Optimization of genome editing by CRISPR ribonucleoprotein for high efficiency of germline transmission of Sox9 in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
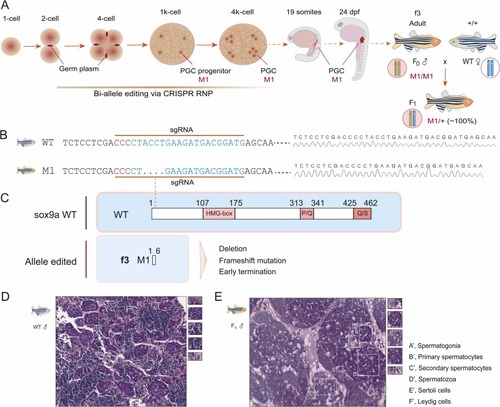
Bi-allele editing, germline transmission, and histology of the edited testis in the sox9a f3 founder. A. Genome editing window during early embryo development, biallelic editing in the first 4 PGC progenitors, and germline transmission. The edited PGCs migrate to their destination to form gonads, in which germ cells have the M1/M1 genotype. M1 indicates the allele 1 edited. The f3 founder crossing with wild-type zebrafish generated all the F1 offspring with the M1/+ genotype. B. DNA sequencing of the edited sox9a gene in the f3 founder and F1 generations. Sequencing diagrams are shown in the right panel. Deletion of 4 bp is indicated in the sgRNA region. The pan sites are denoted in red. C. Schematic diagram of truncated sox9a proteins owing to deletion, frameshift, and early termination compared to the wild type. The HMG box, P/Q, and Q/S domains are denoted in red-shaded boxes. The dotted line indicates the sgRNA site. WT, wild-type. D, E. H&E analysis of the wild-type testis (D) and the sox9a-edited testis in the f3 founder (E). A’, spermatogonia; B’, primary spermatocytes; C’, secondary spermatocytes; D’, spermatozoa; E’, Sertoli cells; F’, Leydig cells. |
Reprinted from New biotechnology, , Yang, K., Cai, L., Zhao, Y., Cheng, H., Zhou, R., Optimization of genome editing by CRISPR ribonucleoprotein for high efficiency of germline transmission of Sox9 in zebrafish, , Copyright (2025) with permission from Elsevier. Full text @ N. Biotechnol.