|
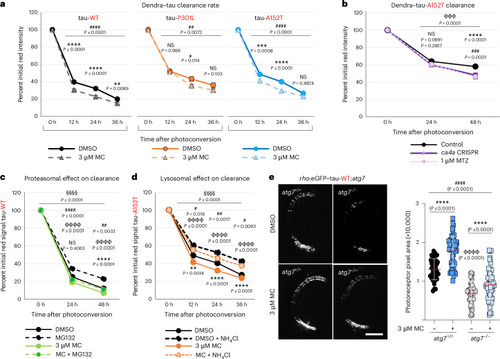
CA inhibition increases the clearance rate of tau without affecting proteasomal or autophagic degradation in vivo. a, Clearance kinetics of Dendra–tau in fish expressing tau-WT, tau-P301L or tau-A152T treated with DMSO (continuous lines) or 3 μM methocarbamol (dashed lines). All Dendra–tau forms cleared at a significantly faster rate following methocarbamol treatment than following treatment with DMSO (n ≥49 neurons per group; data are shown as mean ± s.d.). Data were analyzed by two-way ANOVA (##P ≤ 0.01 and ####P ≤ 0.0001) followed by Sidak’s multiple comparisons, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001 versus DMSO. b, Clearance kinetics of Dendra–tau-A152T in fish treated with 1 μM methazolamide or injected with CRISPR guide RNA targeting ca4a compared with uninjected fish (treated with DMSO). Treatment with methazolamide or ca4a genetic inhibition increased the clearance of Dendra–tau-A152T (n ≥ 57 neurons per group; data are shown as mean ± s.d.). Data were analyzed by one-way ANOVA, ɸɸɸP ≤ 0.001, followed by Sidak’s multiple comparisons test with the following significance values: ****P ≤ 0.0001 versus control for ca4a CRISPR and ###P ≤ 0.001 versus control for methazolamide. c, Clearance kinetics of Dendra–tau-WT in fish treated with 3 μM methocarbamol with or without the proteasome inhibitor MG132 (10 μM). MG132 delayed the clearance of tau-WT, whereas methocarbamol increased tau clearance. However, tau-WT clearance in the presence of methocarbamol + MG132 was significantly higher than that observed in the presence of MG132 alone, suggesting that methocarbamol can accelerate tau clearance when proteasomal degradation is inhibited (n ≥ 69 neurons per group; data are shown as mean ± s.d.). Data were analyzed by one-way ANOVA, §§§§P ≤ 0.0001, followed by Sidak’s multiple comparisons test with the following significance values: ##P < 0.01 and ####P < 0.0001 for DMSO versus methocarbamol; ɸɸɸɸP ≤ 0.0001 for DMSO versus MG132; ****P ≤ 0.0001 for methocarbamol versus methocarbamol + MG132. d, Effect of methocarbamol on lysosomal tau clearance kinetics of Dendra–tau-A152T in the presence or absence of 10 μM NH4Cl. NH4Cl blocks lysosomal acidification and delays the clearance of tau-A152T, whereas 3 μM methocarbamol accelerates it. Tau is cleared more rapidly when methocarbamol is combined with NH4Cl than with NH4Cl alone, suggesting that methocarbamol-improved tau clearance kinetics do not entirely depend on lysosomal degradation (n ≥ 60 neurons per group; data are shown as mean ± s.d.). Data were analyzed by one-way ANOVA, §§§§P ≤ 0.0001, followed by Sidak’s multiple comparisons test with the following significance values: **P ≤ 0.01 and ****P ≤ 0.0001 for DMSO versus 3 μM methocarbamol; #P < 0.05 and ##P < 0.01 for DMSO + NH4Cl versus methocarbamol + NH4Cl; ɸɸɸɸP ≤ 0.0001 for DMSO versus DMSO + NH4Cl. e, Representative images and quantification of photoreceptors in autophagy-competent or autophagy-null rho:eGFP–tau-WT fish. Autophagy abrogation accelerated the loss of rod photoreceptors, whereas 3 μM methocarbamol rescued retinal degeneration in both atg7+/+ and Atg7-deficient (atg7−/−) fish to the same extent; scale bar, 50 μm (n ≥ 39 eyes per condition; data are shown as mean ± s.d.). Data were analyzed by one-way ANOVA, ####P ≤ 0.0001, followed by Tukey’s multiple comparisons test with the following significance values: ****P ≤ 0.0001 versus DMSO (–) and ɸɸɸɸP ≤ 0.0001 versus atg7+/+. Source data
|