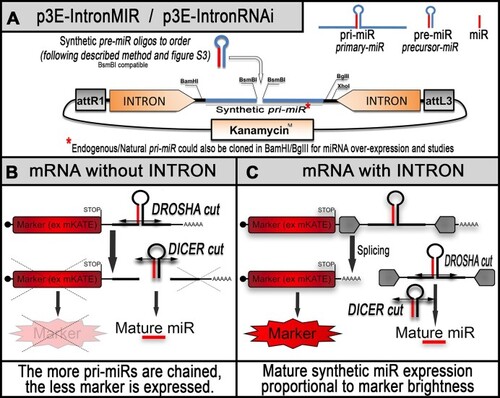
Schematics of the new RNAi backbone for zebrafish gene silencing. (A) Tol2kit compatible p3E presenting an optimized empty synthetic pri-miR (or RNAi cassette) embedded into a β-globin intronic sequence. This presented pri-miR is designed to allow rapid directional insertion of synthetic pre-miR of choice. Synthetic pre-miR(s) of choice are generated by annealing specific top and bottom RNAi oligos designed to release a mature miRNA directed against the 3′UTR of a gene-of-interest [see ‘Methods’ section and previous material (5)]. The RNAi cassette is flanked by restriction sites allowing subsequent and repetitive chaining [see ‘Methods’ and Supplementary Figure S2) for generating p3E-RNAi plasmid with multiple pri-miR for either increasing the potency of the desired knockdown or targeting multiple genes at the same time. (B) Without intron, the pre-miR/RNAi cassette is transcribed along with a co-marker on the same RNA, which is cut by Drosha in the nucleus for releasing the associated pre-miR. This cut leaves the mRNA without polyA tail and leads to its rapid degradation. The associated fluorescence is not a good indicator of the activity and amount of synthetic miRNA produced. (C) The presence of the intronic sequence is designed to rescue the co-expression of the marker (see also Supplementary Figure S1).
|

