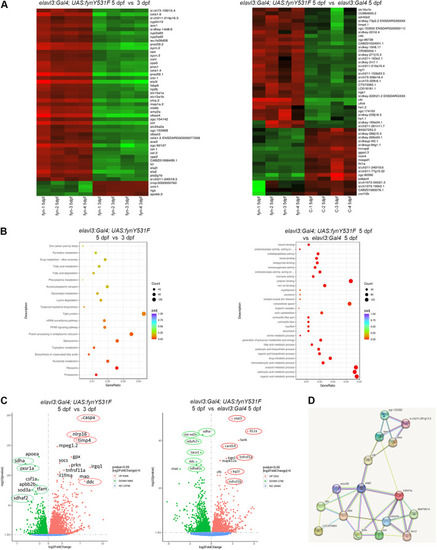
Transcriptome analysis reveals that FynY531F signaling alters neuroprotective, oxidative and metabolic pathways, and identifies Stat3 as a downstream Fyn effector. (A) Heatmaps of highest gene expression changes comparing bulk RNA sequencing from 5 dpf versus 3 dpf elavl3:Gal4; UAS:fynY531F larvae (n=4; left), and 5 dpf elavl3:Gal4; UAS:fynY531F versus 5 dpf elavl3:Gal4 larvae (n=4; right). (B) Pathway analysis reveals altered metabolic pathways in 5 dpf versus 3 dpf elavl3:Gal4; UAS: fynY531F larvae (left) and 5 dpf elavl3:Gal4; UAS:fynY531F versus 5 dpf elavl3:Gal4 larvae (right). (C) Volcano plots showing upregulated and downregulated genes in 5 dpf versus 3 dpf elavl3:Gal4; UAS:fynY531F larvae (left) and 5 dpf elavl3:Gal4; UAS:fynY531F versus 5 dpf elavl3:Gal4 larvae (right). Nuclear-encoded mitochondrial genes sdha, sdhaf2 and taco1 are reduced. Elevated genes include stat3, caspa, tngrsf1a/b and irg1l. (D) STRING network analysis identifies Fyn interactions with Tnf-α and Stat3 signaling pathways in elavl3:Gal4; UAS:fynY531F larval transcriptome.
|