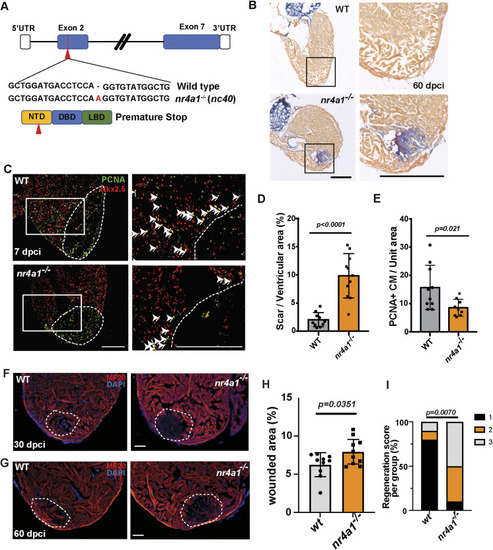
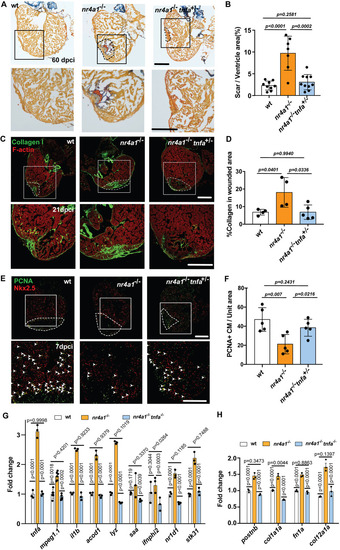
Persistent fibrosis in nr4a1 mutant after cardiac injury. (A-C) Left: distribution of fibroblasts on cardiac sections of uninjured wild-type (WT) hearts or WT hearts at 21 and 30 dpci. Right: distribution of fibroblasts on cardiac sections of uninjured nr4a1 mutant hearts or nr4a1 mutant hearts at 21 and 30 dpci. (D,E) Quantification of fibroblasts per unit area for WT and nr4a1 mutant hearts at 21 and 30 dpci. (F,H) Collagen deposition, as revealed by collagen I antibody staining, in WT and nr4a1 mutant at 21 dpci and 30 dpci, respectively. (G,I) Quantification of collagen deposition in the wounded area of WT and nr4a1 mutant hearts at 21 dpci and 30 dpci, respectively. (J,L) α-SMA immunostaining at 30 dpci and 60 dpci injured hearts, respectively. Left: WT group; Right: nr4a1 mutant group. (K,M) Quantification of α-SMA signal in the wounded area at 30 dpci and 60 dpci, respectively. (N) Schematic showing persistent fibrosis in nr4a1 mutant. Dashed lines show the wounded area. Boxed regions show the approximate positions for quantification, and position of magnified images. Created in BioRender by Feng, D., 2025. https://BioRender.com/ok17j9e. This figure was sublicensed under CC-BY 4.0 terms. P-values<0.05 were considered statistically significant (two-way ANOVA with Sidak test for multiple comparison correction and two-tailed unpaired t-test). Points in D, E, G, I, K and M show the individual samples. Data are mean±s.e.m. Scale bars: 275 µm.
|