- Title
-
The USH3A causative gene clarin1 functions in Müller glia to maintain retinal photoreceptors
- Authors
- Nonarath, H.J.T., Simpson, S.L., Slobodianuk, T.L., Tran, H., Collery, R.F., Dinculescu, A., Link, B.A.
- Source
- Full text @ PLoS Genet.
|
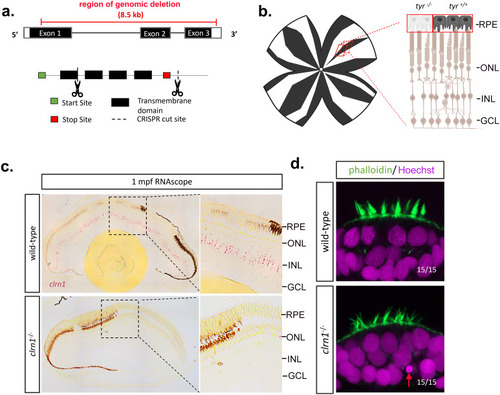
Development of USH3A zebrafish model. CRISPR/Cas9 technology was used to generate a large deletion within the |
|
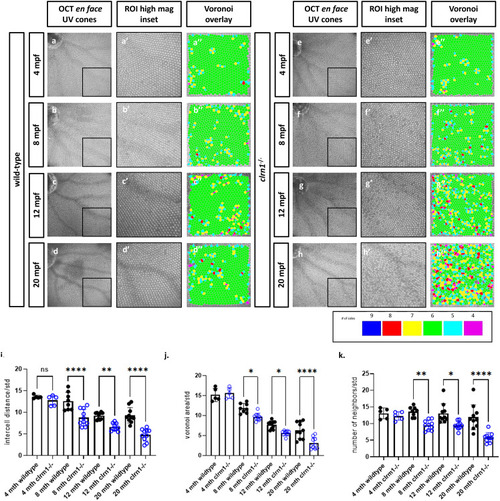
clrn1-/- zebrafish present with an altered UV cone mosaic beginning at 8 mpf. |
|
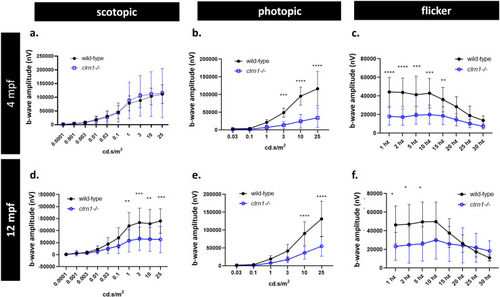
Scotopic and photopic ERG responses are affected by the loss of Clrn1. Quantification of 4 mpf wild-type and |
|
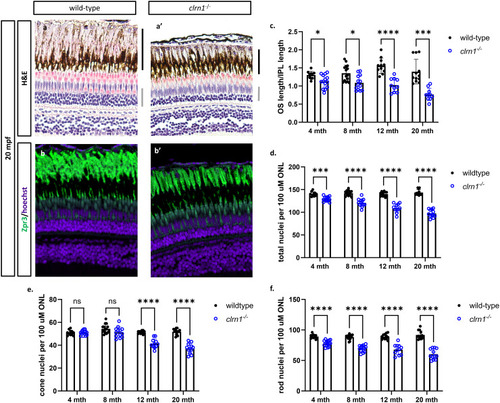
clrn1-/- zebrafish present with age-dependent shortening of the rod outer segments and thinning of the outer nuclear layer. Hematoxylin and eosin-stained paraffin section from (a) 20 mpf wild-type and (a’) |
|
High-intensity light induces rod and cone cell death in (a) Overview of light stress assay: larvae were treated with 3500 lux for 48 hours from 5-7 dpf. (b) Representative images of TUNEL-positive photoreceptors (white arrows) on cryosectioned retina from 7 dpf wild-type (upper panels) and |
|
Actin is disorganized in the outer retina of Transverse sections from 7 dpf wild-type and |
|
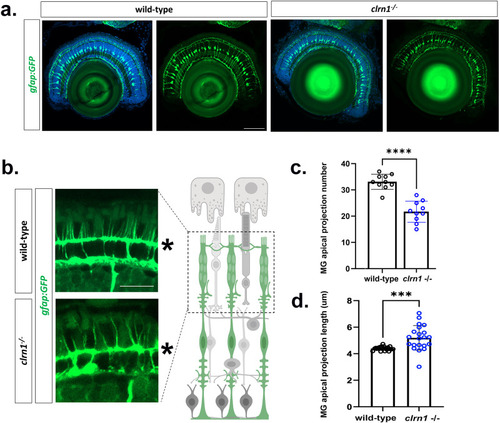
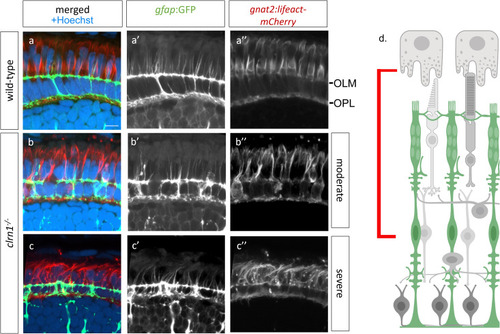
Apical processes of Müller glia and calyceal processes of cone photoreceptors are disorganized in Transverse sections from 7 dpf (a) wild-type and (b,c) |
|
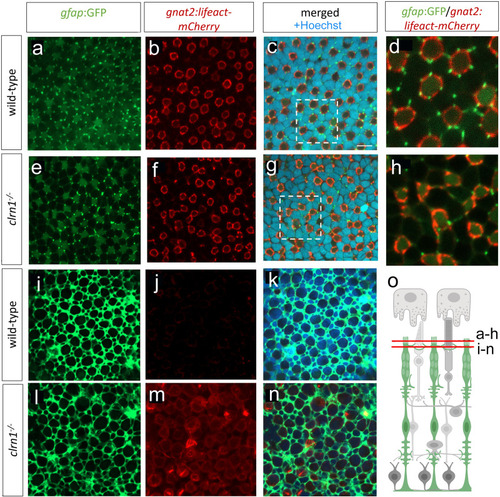
Interactions between Müller glia and cone photoreceptor cells are disorganized in |
|
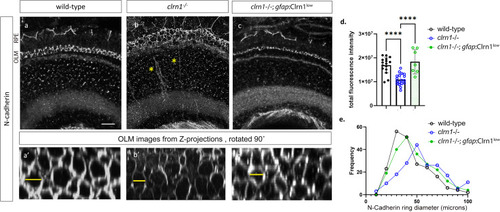
Expression of N-cadherin is reduced in Confocal microscope images of N-cadherin immunostaining in transverse cryosections of retinas from 7 dpf (a) wild-type, (b) |
|
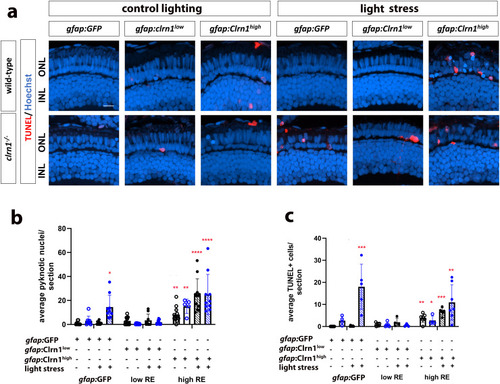
Therapeutic efficacy of Clrn1 re-expression in Müller glia depends on expression level. (a) Retinal images of 7 dpf wild-type (upper rows) and |
|
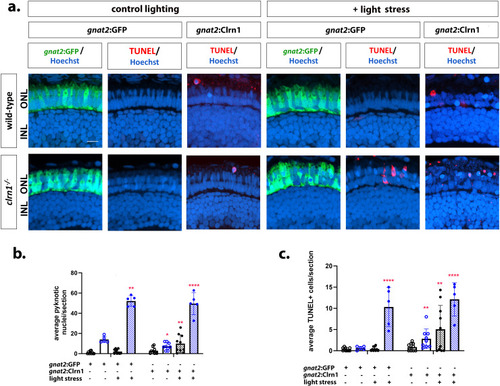
Re-expression of Clrn1 in cone photoreceptors does not reduce sensitive to light stress. (a) Retinal images of 7 dpf wild-type (upper rows) and |
|
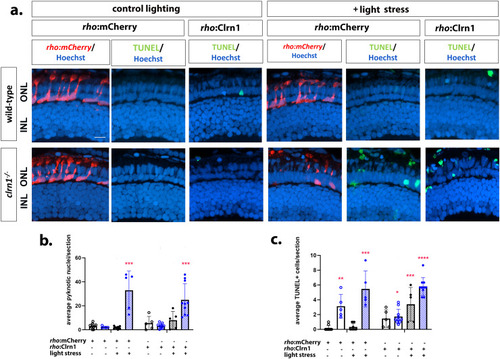
Re-expression of Clrn1 in rod photoreceptors does not reduce sensitive to light stress. (a) Retinal images of 7 dpf wild-type (upper rows) and |