- Title
-
ZebraReg-a novel platform for discovering regulators of cardiac regeneration using zebrafish
- Authors
- Apolínová, K., Pérez, F.A., Dyballa, S., Coppe, B., Mercader Huber, N., Terriente, J., Di Donato, V.
- Source
- Full text @ Front Cell Dev Biol
|
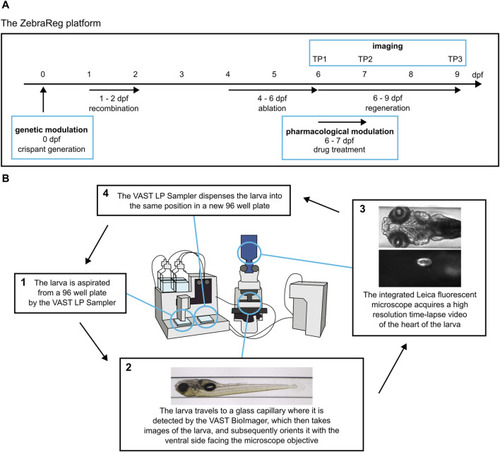
ZebraReg: a novel platform for discovering regulators of cardiac regeneration using zebrafish. |
|
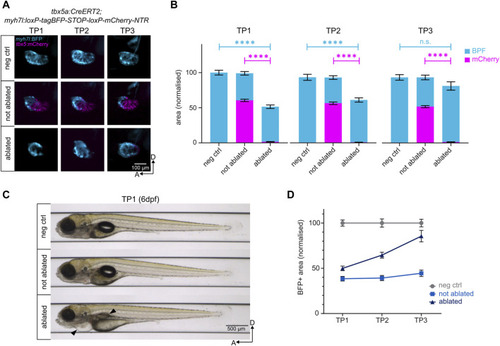
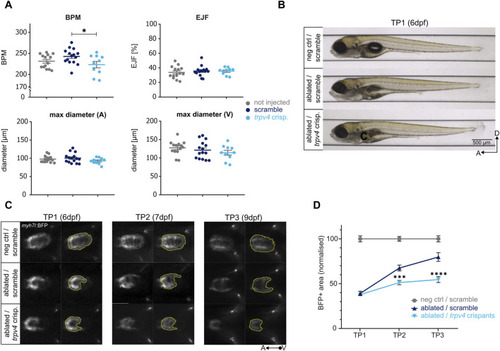
Genetic ablation of ventricular cardiomyocytes and subsequent regeneration. |
|
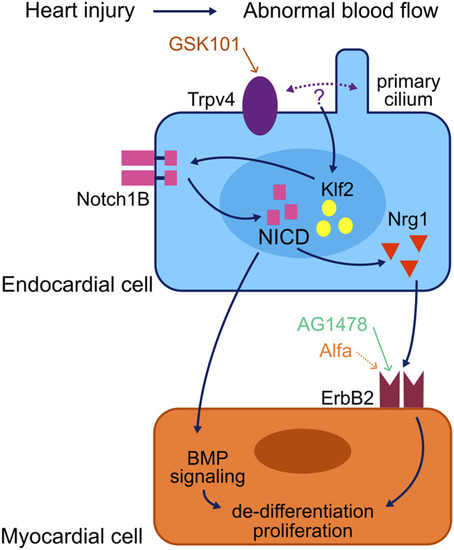
Model of a myocardial regeneration signalling network. Activation of the mechanosensory channel Trpv4 in the endocardium triggers a series of signalling events which lead to the activation of the ErbB2 channel in the myocardium and thus the initiation of regenerative processes in the injured myocardium. Adapted from |
|
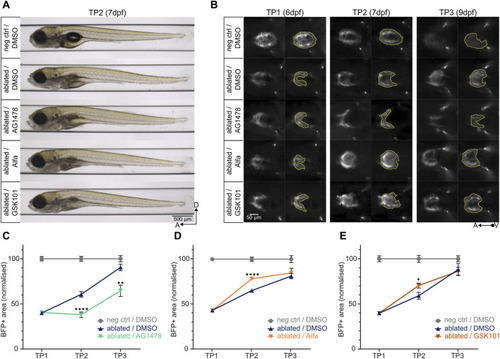
Determining the pro- and anti-regenerative effects of drugs on regeneration kinetics. |
|
Determining the effect of genes on regeneration kinetics. The CRISPR/Cas9-driven loss of function of |