Fig. 1
- ID
- ZDB-FIG-240626-44
- Publication
- Silvius et al., 2024 - A Robust Closed-Tube Method for Resolving tp53M214K Genotypes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
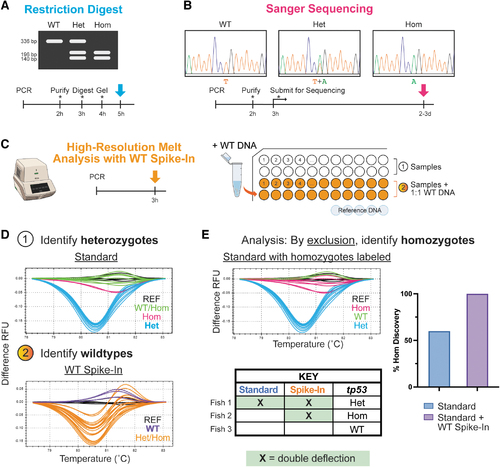
An improved workflow for tp53M214K genotyping. Conventional genotyping protocols for the tp53M214K mutant, including (A) restriction digest and (B) Sanger sequencing-based approaches are time-consuming and/or require multiple hands-on steps (indicated by asterisk), whereas our protocol uses (C) HRMA and requires a single PCR program on a standard qPCR machine (left); samples are run in parallel with and without WT DNA spiked in at a 1:1 ratio alongside a set of WT reference DNA for comparison (right). (D) The analysis strategy utilizes the characteristic deflection pattern of tp53 heterozygotes to first identify heterozygotes in the standard run with 1 μL sample DNA; putative homozygotes that can be called with the standard HRMA are indicated in pink (1). Since WT DNA added to homozygotes yields a similar pattern to heterozygotes, samples that do not demonstrate that deflection pattern with the spike-in can be identified as WTs (2). (E) True homozygotes can then be identified by comparative exclusion as illustrated in the table. The graph shows all homozygotes (also verified by sequencing) in pink overlaid on the standard curve. When only the standard samples are evaluated, only ∼60% of homozygotes are called. However, when the parallel spike-in is analyzed, all homozygotes are correctly identified. Het, heterozygote; Hom, homozygote; HRMA, high-resolution melt analysis; qPCR, quantitative polymerase chain reaction; REF, reference DNA; RFU, relative fluorescence units; WT, wild-type. |