|
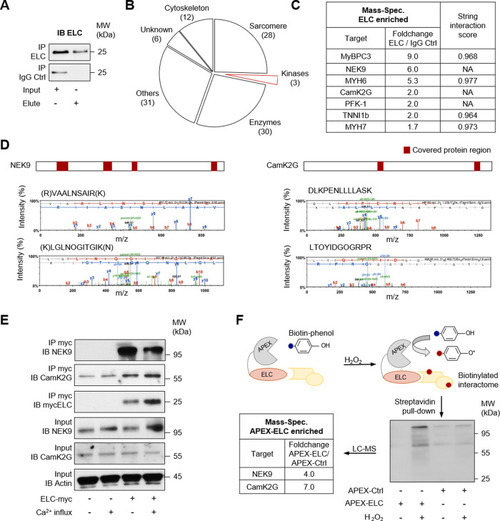
Identification of ELC kinases in adult piscine heart protein.Immunoprecipitation (IP) was performed by using custom ELC antibodies covalently coupled to magnetic beads. Liquid chromatography followed by mass spectrometry (LC-MS) was used for identification. A Validation of ELC precipitation by immunoblot (IB) before performing LC-MS (see B and C). B Classification of ELC enriched proteins identified by LC-MS. From 3452 spectra 110 proteins were identified in total. 106 proteins were found to interact with ELC. 3 different kinases were found. (Peptide FDR: 5%, minimum of two identified peptides per protein) C Ranking of ELC enriched proteins by foldchange of unique peptide counts enriched by ELC compared to unique peptide counts of the same protein captured by the isotype control (IgG Ctrl). Data of the screening experiment are shown without statistical analysis. Candidates were validated using independent methods. The string intersection score was calculated for known ELC interacting proteins67. (NA: not available) D Two representative mass spectrometry spectra of unique peptides accepted to identify NIMA related kinase 9 (NEK9) (left panel) or calcium/calmodulin-dependent protein kinase type II subunit gamma (CamK2G) (right panel). The protein coverage is shown above (red boxes). E Validation of ELC-kinase interaction. Human myc-tagged ELC protein was overexpressed in human cells followed by specific immunoprecipitation (IP) and immunoblot (IB) analysis. Ca2+-influx was obtained by ionophore stimulation. One representative experiment is shown (see also Fig. 2D). F Schematic illustration of ascorbate peroxidase (APEX) catalyzed proximity labeling. Overexpression of ELC fused to APEX enables biotin-labeling of proteins in close proximity. The reaction is catalyzed by hydrogen peroxide (H2O2). Streptavidin precipitation followed by LC-MS analysis identified NEK9 and CamK2G. The foldchange of unique peptide counts of biotin-labeled proteins in the proximity of ELC (APEX-ELC) compared to unique peptide counts of the same protein in the APEX-Ctrl sample is shown. Data of the validation experiment are shown without statistical analysis.
|