- Title
-
Forward genetic screen in zebrafish identifies new fungal regulators that limit host-protective Candida-innate immune interaction
- Authors
- Blair, B.A., Bragdon, E., Dhillon, G., Baker, N., Stasiak, L., Muthig, M., Miramon, P., Lorenz, M.C., Wheeler, R.T.
- Source
- Full text @ MBio
|
Defining infection parameters. (A) Flow chart showing the workflow of pilot experiments. Hindbrain infections were performed at the prim-25 stage, and fish were then screened to ensure they received the correct inoculum (10–25 cells). At 4–6 hours postinfection, fish were imaged by confocal microscopy to score fungal phagocytosis; survival was monitored out to 72 hpi. (B) Example Kaplan-Meier survival curves pooled from three experiments showing the fish injected with PBS (control, |
|
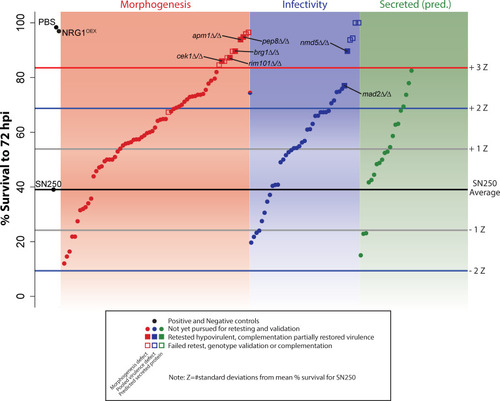
High-throughput virulence screening. Average survival of fish infected with individual mutant |
|
Complementation restores virulence to hypovirulent |
|
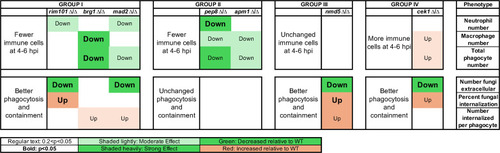
Mutants grouped by altered innate immune response. Mutant–phagocyte interactions were scored at 4–6 hpi, relative to wild-type fungi–phagocyte interactions in the same experiment, as to immune cell number and phagocytosis efficiency traits. Group I mutants had lower numbers of recruited phagocytes and better phagocytosis and containment. Group II mutants had fewer recruited phagocytes and unchanged phagocytosis efficiency. The Group III mutant had no change in phagocyte recruitment but better phagocytosis. The Group IV mutant had higher phagocyte recruitment and better phagocytosis. “Down” or “up” text indicates the direction of differential immune response, bold lettering indicates |
|
Phagocyte recruitment to hypovirulent |
|
Phagocytosis of hypovirulent |
|
Fungal morphology at 4–6 hpi. Mutant fungal morphology was scored at 4–6 hpi, relative to wild-type fungal morphology in the same experiment. Two of the three Group I mutants and both the Group II mutants had decreased filamentous growth at 4–6 hpi. “Down” text indicates decreased filamentous morphology (pseudohyphal/hyphal). Bold lettering indicates |
|
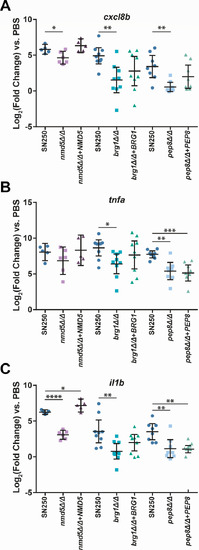
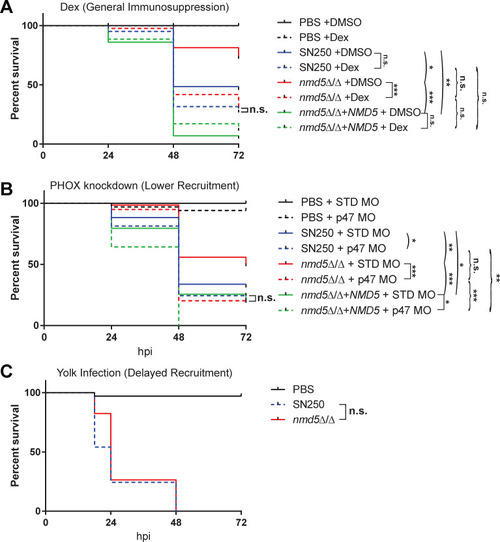
Hypovirulent |
|
|