- Title
-
Rapid and robust generation of cardiomyocyte-specific crispants in zebrafish using the cardiodeleter system
- Authors
- Keeley, S., Fernández-Lajarín, M., Bergemann, D., John, N., Parrott, L., Andrea, B.E., González-Rosa, J.M.
- Source
- Full text @ Cell Rep Methods
|
The cardiodeleter transgene efficiently creates cardiomyocyte-specific LOF phenotypes in combination with gene-specific gRNAs (A) Schematic representation of the cardiodeleter transgene, which drives the expression of nGFP and a zebrafish codon optimized version of Cas9 in cardiomyocytes. (B and B′) Representative heart from an adult zebrafish carrying the cardiodeleter allele, exhibiting normal morphology (B) and expression of nGFP (B′). (C) Representation of the ect2 locus in the zebrafish genome. Filled boxes represent coding exons, empty boxes indicate the 5′ UTR and 3′ UTR. Asterisks indicate the location of the three selected gRNA targets, predicted using CRISPRScan. Blue areas indicate the target sequence. Sequences in bold highlight the PAM. (D) Strategy to generate a guide shuttle carrying three U6 promoters (U6a, U6b, and U6c), each driving the expression of a gRNA targeting a GOI, and a transgenesis reporter. (E) Representation of the stable ect2 guide shuttle transgene. (F and F′) Representative heart from an adult zebrafish carrying the ect2 guide shuttle exhibiting normal morphology (F) and expression of n-mKate (F′). (G and H) The 30-dpf cardiodeleter+ (G) and cardiodeleter+ect2 guide shuttle+ (H) animals. Stalled growth and edema can be observed only in double transgenic animals. (Right) Dissected hearts. (I and J) Confocal images of DAPI-stained whole hearts from 30-dpf cardiodeleter+ (K) and cardiodeleter+ect2 guide shuttle+ (L) animals. Boxed regions show DAPI staining at higher magnification (white). Asterisks: polyploid nuclei. Cyan and yellow arrowheads: non-myocytes and cardiomyocytes, respectively. AT, atrium; BA, bulbus arteriosus; Chr, chromosome; V, ventricle. Scale bars, 200 μm (B and F) and 20 μm (I and J). |
|
An orthogonal transposase approach is required to efficiently generate cardiomyocyte-specific crispants (A) Representation of the tnnt2a locus in the zebrafish genome, indicating the exons targeted by the selected gRNAs (asterisks). (B–E) Confocal projections of representative 96 h post fertilization (hpf) embryonic hearts from wild-type (B and C) or cardiodeleter+ animals (D and E) from the indicated experimental groups, immunostained to detect cardiomyocyte nuclei (white) and Tnnt2a (cyan). The percentage of Tnnt2a-negative (Tnnt2a–) cardiomyocytes is summarized in the bottom left corner (average ± SD, n = 4, 7, 6, and 8 embryos). (F and J) Confocal projections of 96 hpf embryonic hearts from animals carrying the cardiodeleter transgene and injected with a Tol2- (F; n = 8) or Tol1- (J; n = 7) based guide shuttle encoding gRNAs targeting tnnt2a, immunostained to detect Tnnt2a (cyan). Yellow and white arrowheads indicate Tnnt2a+ and Tnnt2a– guide shuttle+ cardiomyocytes (magenta), respectively. Single confocal planes are shown in F′ and J′. (G–I) Representative ventricular sections from ∼30 dpf cardiodeleter+ animals, uninjected (G) or injected with the indicated combinations of guide shuttles and mRNA (H and I). White arrows indicate cardiomyocyte nuclei. Yellow arrows and cyan arrowheads indicate mKate+cardiodeleter+ and mKate+cardiodeleter– cardiomyocytes, respectively. Cartoons (right) summarize the observed patterns of excision and integration of the cardiodeleter and guide shuttles. Chr, chromosome. Scale bars, 20 μm. |
|
Cardiodeleter-induced crispants carry biallelic mutations in ∼90% of labeled cardiomyocytes (A) Representation of the location of the gRNAs targeting tnnt2a, cmlc2, and amhc. Tol1-guide shuttles encoding three guides for each gene were injected into wild-type or cardiodeleter+ embryos at the one-cell stage. Embryos exhibiting mKate+ clones in the heart, indicative of guide shuttle integration, were selected at 4 dpf and fixed for immunostaining. (B and C) Confocal projections of representative 96 h post fertilization embryonic hearts from wild-type (B) or cardiodeleter+ animals (C) injected with guide shuttles targeting the indicated genes, immunostained with specific antibodies to recognize the corresponding protein (cyan). Cyan arrowheads indicate guide shuttle (mKate)+ cardiomyocytes with biallelic mutations, leading to the complete loss of the corresponding protein. Yellow arrows indicate mKate+ cardiomyocytes maintaining protein expression. (D) Quantification of the efficiency of biallelic mutations in wild-type or cardiodeleter embryos injected with the indicated guide shuttles. Individual values plotted; black line: average. p values: two-tailed unpaired t test. AT, atrium; chr, chromosome; V, ventricle. Scale bars, 20 μm. |
|
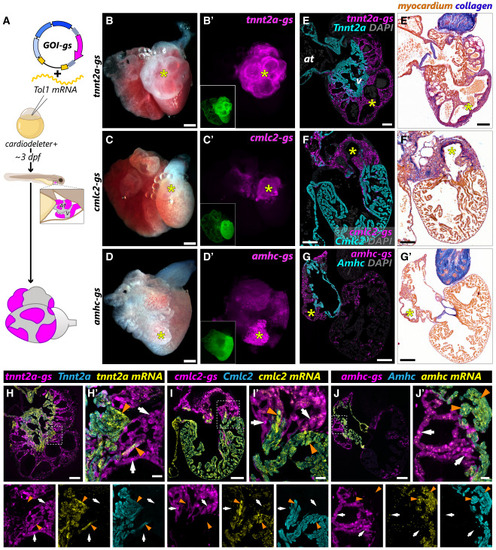
Cardiomyocyte clones carrying biallelic mutations in tnnt2a, cmlc2, or amhc survive to adulthood intermingled with wild-type cells (A) Schematic representation of the experimental approach to test whether labeled myocardial clones carrying biallelic mutations can be recovered in adult hearts. (B–D) Representative images of dissected hearts from cardiodeleter+ animals injected with the indicated guide shuttles. (B′–D′) mKate signal from hearts shown in (B–D), indicating cardiomyocyte clones that have incorporated the corresponding guide shuttle. The GFP fluorescence from the cardiodeleter transgene is shown in the images located in the bottom left corner. (E–G) Histological sections from hearts shown in (B–D), immunostained with antibodies to identify guide shuttle+ cardiomyocytes (mKate+, magenta) and the corresponding protein encoded by the targeted gene (cyan). (E′–G′) Consecutive sections from hearts shown in (E–G) stained using the AFOG technique to identify collagen (blue) and myocardium (orange/brown). Yellow asterisks indicate large mKate+ clones that show complete absence of the corresponding protein. (H–J) Combined immunofluorescence-RNAScope in consecutive sections from hearts shown in (E–G). Signal from mRNA ISH appears in yellow. White arrows indicate guide shuttle (mKate)+ cardiomyocytes with biallelic mutations, leading to the reduction of the corresponding mRNA and the complete loss of the corresponding protein. Orange arrowheads point to guide shuttle– cardiomyocytes that maintain high levels of mRNA and protein expression. at: atrium; v, ventricle. Scale bars, 200 μm (B–D), 100 μm (E–I and E′–G′), and 20 μm (H′–J′). |
|
A guide shuttle encoding multiple gRNAs targeting amhc is required for highly efficient biallelic mutagenesis (A–C) Confocal projections of representative 96 h post fertilization embryonic hearts from cardiodeleter+ animals injected with guide shuttles carrying three (A), two (B), or one (C) gRNAs targeting amhc. (D) Quantification of the efficiency of biallelic mutations in wild-type and cardiodeleter embryos injected with the indicated guide shuttles. Individual values plotted; black line: average. p values: one-way ANOVA, followed by Tukey’s multiple comparisons test. (E–G) Histological sections of hearts from adult cardiodeleter animals injected with the indicated guide shuttle, immunostained with antibodies to identify mKate (guide shuttle, magenta) and Amhc (cyan). Amhc signal is shown in (E′–G′). Cyan arrowheads indicate mKate (guide shuttle)+ cardiomyocytes with biallelic mutations that lead to complete depletion of the Amhc protein. Orange arrows indicate guide shuttle+ cardiomyocytes that maintain detectable levels of Amhc expression. (H) Efficiency of biallelic mutations in wild-type and cardiodeleter adult animals injected with the indicated guide shuttles. Individual values plotted; black line: average. p values: one-way ANOVA, followed by Tukey’s multiple comparisons test. (I) Distribution of guide shuttle+ cardiomyocyte population according to Amhc protein levels in wild-type and cardiodeleter adult animals injected with the indicated guide shuttles. at, atrium; v, ventricle. Scale bars, 20 μm (A–C) and 100 μm (E–G). |
|
Long-term tracking of erbb2 myocardial crispants and stable-myocardial mutants demonstrates a cell-autonomous role in trabeculation (A) Representation of the location of the gRNAs targeting erbb2. (B and C) Representative single plane confocal images of embryonic hearts at 4 dpf from wild-type (B) and cardiodeleter+ animals (C), injected with a guide shuttle encoding gRNAs targeting erbb2 (erbb2-gs), immunostained to detect Tnnt2a (cyan). (D and E) Representative heart sections from animals of the indicated genotypes, injected with the erbb2-gs, analyzed at 40 dpf. Magnifications of the boxed areas in D and E are shown in (D′–E′′). Yellow arrowheads, guide shuttle+ cardiomyocytes located at the primordial myocardium; white arrows, guide shuttle+ trabecular cardiomyocytes; asterisks, trabecules. (F and G) Representative images of 4 dpf embryonic hearts from the indicated genotypes, immunostained to detect nGFP (cardiodeleter, green) and mKate (erbb2-gs, magenta). Cyan arrowheads, primordial cardiomyocytes; white arrows, trabecular myocardium. (H and I) Representative sections of cardiodeleter+ (H) and cardiodeleter+erbb2-gs+ siblings (I) stained with the AFOG technique to detect collagen (blue) and myocardium (orange/brown). Whole fish pictures shown on the top, right corner. (H′ and I′) Consecutive sections from the hearts shown in (H) and (I), immunostained to detect nGFP (cardiodeleter, green) and mKate (erbb2-gs, magenta). at, atrium; v, ventricle. Scale bars, 20 μm (B, C, F, and G), 100 μm (D, E, H, and I). |