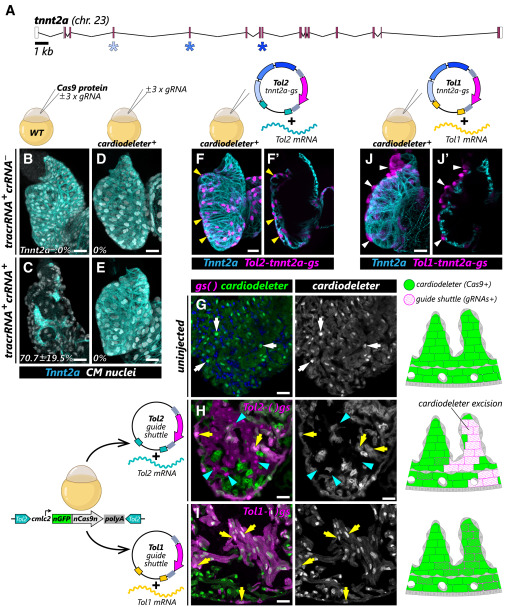
Fig. 2 An orthogonal transposase approach is required to efficiently generate cardiomyocyte-specific crispants (A) Representation of the tnnt2a locus in the zebrafish genome, indicating the exons targeted by the selected gRNAs (asterisks). (B–E) Confocal projections of representative 96 h post fertilization (hpf) embryonic hearts from wild-type (B and C) or cardiodeleter+ animals (D and E) from the indicated experimental groups, immunostained to detect cardiomyocyte nuclei (white) and Tnnt2a (cyan). The percentage of Tnnt2a-negative (Tnnt2a–) cardiomyocytes is summarized in the bottom left corner (average ± SD, n = 4, 7, 6, and 8 embryos). (F and J) Confocal projections of 96 hpf embryonic hearts from animals carrying the cardiodeleter transgene and injected with a Tol2- (F; n = 8) or Tol1- (J; n = 7) based guide shuttle encoding gRNAs targeting tnnt2a, immunostained to detect Tnnt2a (cyan). Yellow and white arrowheads indicate Tnnt2a+ and Tnnt2a– guide shuttle+ cardiomyocytes (magenta), respectively. Single confocal planes are shown in F′ and J′. (G–I) Representative ventricular sections from ∼30 dpf cardiodeleter+ animals, uninjected (G) or injected with the indicated combinations of guide shuttles and mRNA (H and I). White arrows indicate cardiomyocyte nuclei. Yellow arrows and cyan arrowheads indicate mKate+cardiodeleter+ and mKate+cardiodeleter– cardiomyocytes, respectively. Cartoons (right) summarize the observed patterns of excision and integration of the cardiodeleter and guide shuttles. Chr, chromosome. Scale bars, 20 μm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep Methods