- Title
-
Dock1 functions in Schwann cells to regulate development, maintenance, and repair
- Authors
- Doan, R.A., Monk, K.R.
- Source
- Full text @ J. Cell Biol.
|
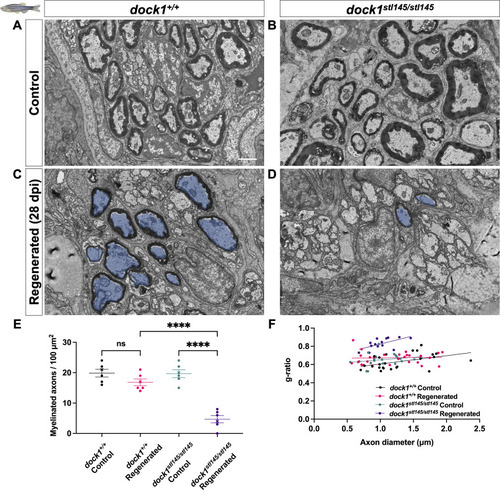
PHENOTYPE:
|
|
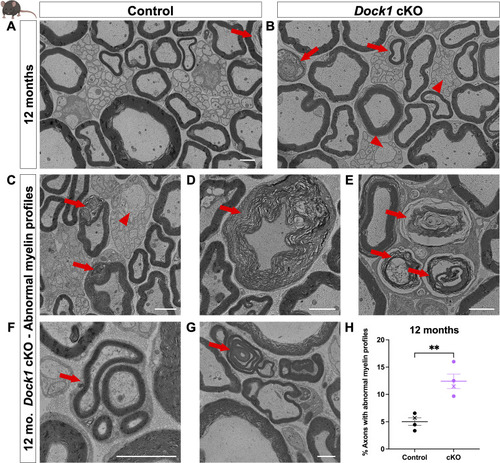
PHENOTYPE:
|
|
|
|
|
|
|
|
|
|
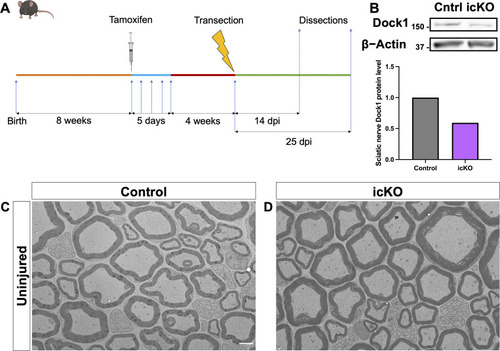
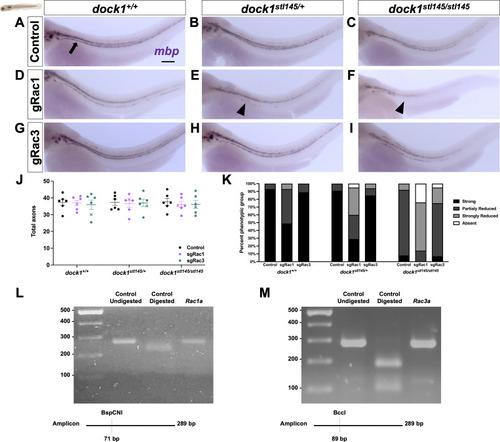
PHENOTYPE:
|
|
|
|
|
|
|
|
|
|
|