- Title
-
Role of ZFHX4 in orofacial clefting based on human genetic data and zebrafish models
- Authors
- Ishorst, N., Hölzel, S., Greve, C., Yilmaz, Ö., Lindenberg, T., Lambertz, J., Drichel, D., Zametica, B., Mingardo, E., Kalanithy, J.C., Channab, K., Kibris, D., Henne, S., Degenhardt, F., Siewert, A., Dixon, M., Kruse, T., Ongkosuwito, E., Girisha, K.M., Pande, S., Nowak, S., Hagelueken, G., Geyer, M., Carels, C., van Rooij, I.A.L.M., Ludwig, K.U., Odermatt, B., Mangold, E.
- Source
- Full text @ Eur. J. Hum. Genet.
|
Workflow of the CNV study and additional individuals/families with |
|
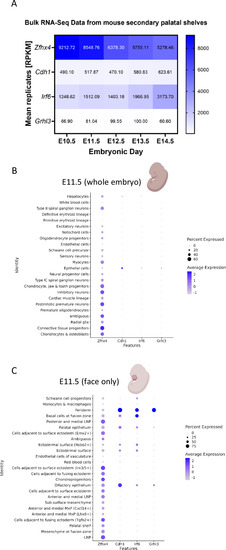
Expression pattern of |
|
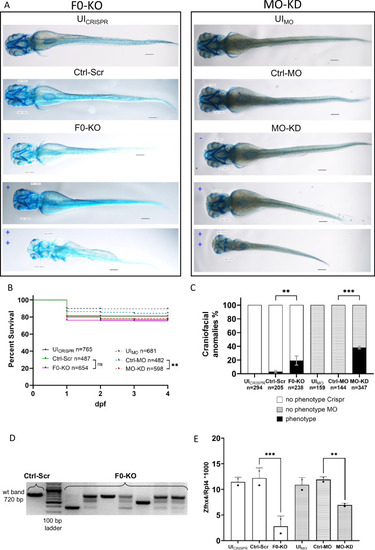
Effect of F0-generation knock-out (F0-KO) and translation-blocking morpholino knock-down (MO-KD) of EXPRESSION / LABELING:
PHENOTYPE:
|
|
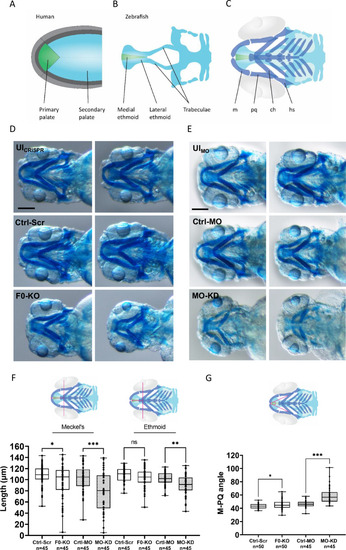
Craniofacial outcomes of PHENOTYPE:
|