- Title
-
Transient receptor potential a1b regulates primordial germ cell numbers and sex differentiation in developing zebrafish
- Authors
- Gong, X., Yan, Q., Chen, L.
- Source
- Full text @ J. Fish Biol.
|
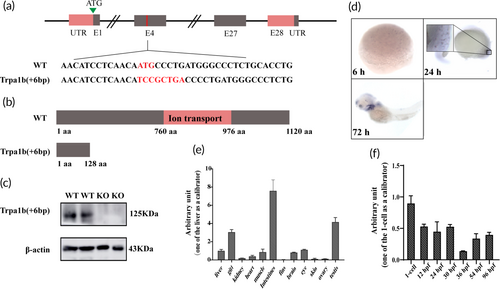
Targeted disruption of the zebrafish trpa1b gene using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9. (a) Structure of the zebrafish trpa1b gene and target site. The mutant site contains a − 3 bp (ATG) DNA sequence and a + 9 bp (TCCGCTGAC) insertion sequence, which includes a stop codon. (b) Schematic representation of the wild-type (WT) and Trpa1b proteins. Functional domains are indicated in red and consist of amino acids (aa) from 760 to 976, which do not reach the ion channel domain. (c) Detection of Trpa1b protein expression at 60 h postfertilization (hpf). KO: Trpa1b−/−. (d) Whole-embryo in situ hybridization showing the temporal expression of trpa1b. (e–f) Analyse the relative expression levels of trpa1b messenger RNA (mRNA) in different adult tissues and embryonic development stages using real-time quantitative PCR (RT-qPCR). The data are presented as mean ± SE of mean (n = 3, biological replicates). |
|
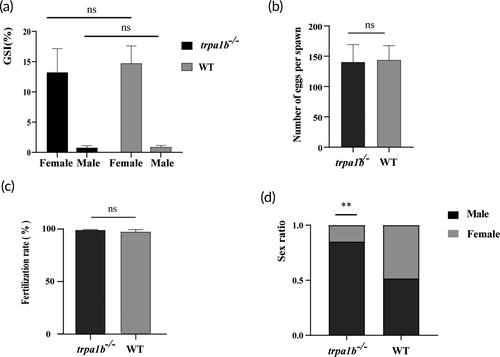
Impact of trpa1b gene disruption on sex ratio in zebrafish. (a) Gonadosomatic index (GSI) of wild-type (WT) and trpa1b−/− zebrafish (n = 6). (b) Ovulation rate of female zebrafish in WT and trpa1b−/− groups (n = 6). (c) Fertilization rate in intercrosses of trpa1b−/− zebrafish (n = 24). (d) Adult zebrafish sex ratio in WT (N = 240) and trpa1b−/− (N = 280) groups. **p < 0.01. PHENOTYPE:
|
|
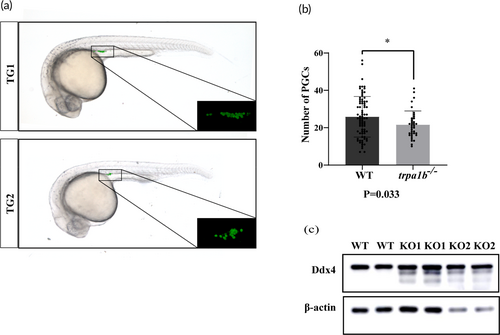
Mutation of the trpa1b gene affects the number of primordial germ cells (PGCs). (a) Distribution of PGCs in zebrafish with different genotypes. TG1, ihb327Tg. TG2, homozygous mutant offspring of trpa1b−/− crossed with TG1. (b) Quantification of PGC numbers in different strains. The numbers of samples were 27 and 56 in the TG1 and TG2 lines, respectively. *p < 0.05. (c) Protein expression of Ddx4 in testis of 6mpf zebrafish. KO1, germline tagged trpa1b−/− strains. KO2, germline unlabeled trpa1b−/− strains. |
|
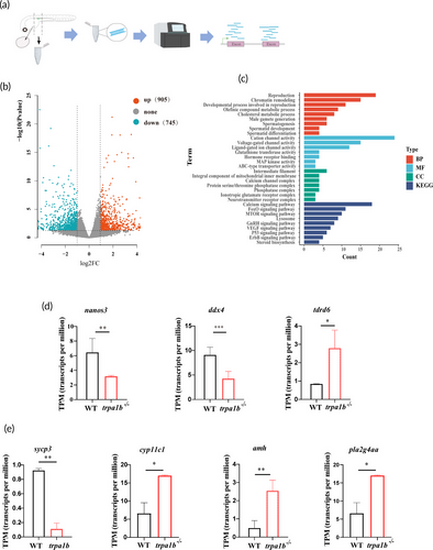
Analysis of differentially expressed genes (DEGs) in Danio rerio. (a) Schematic representation of tissue sampling near the genital ridge in zebrafish at 1.25 days fertilization (dpf) for RNA sequencing (RNA-seq) analysis. (b) Volcano plot depicting DEGs in wild-type (WT) and trpa1b homozygous zebrafish at 1.25 dpf, with 905 upregulated DEGs and 745 downregulated DEGs. (c) Gene ontology (GO) term and KEGG pathway analysis of DEGs in the vicinity of the genital ridge in WT and trpa1b−/− zebrafish, annotated for biological processes (in red), molecular function (in blue), and cellular components (in green) with a p-value cutoff of 0.05. (d) At 1.25 dpf, there is low expression of PGC formation-related genes nanos3 and ddx4, and high expression of the tdrd6 gene. (e) At 1.25 dpf, there is low expression of genes related to spermatid development and differentiation, such as sycp3, and high expression of cyp11c1, amh, and pla2g4aa. *p < 0.05; **p < 0.01; ***p < 0.001. |
|
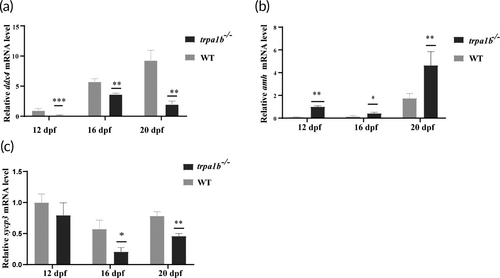
The relative expression of marker genes of germ cell and gonadal somatic cell at 12, 16, and 20 dpf in wild-type (WT) and trpa1b−/− zebrafish (n = 12). (a–c) The relative messenger RNA (mRNA) levels of ddx4, amh, and sycp3. *p < 0.05; **p < 0.01; ***p < 0.0001. EXPRESSION / LABELING:
PHENOTYPE:
|