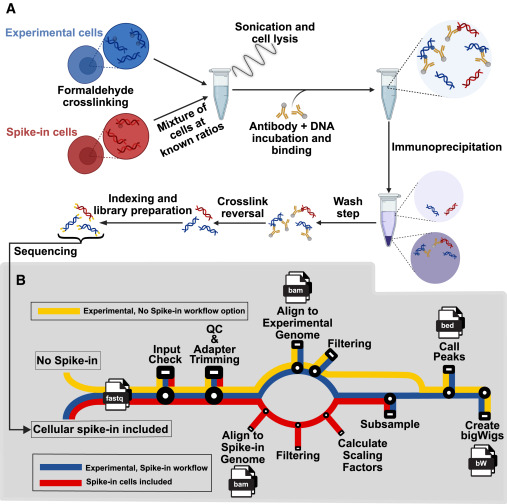
Fig. 2 PerCell workflow (A) Workflow summary for the PerCell spike-in ChIP-seq wet lab procedure. Experimental cells (blue) and spike-in cells (red) of orthogonal species are separately crosslinked with chromatin-associated proteins via formaldehyde fixation before being mixed at known ratios that remain consistent for each sample. Experimental and spike-in cells proceed together throughout the remainder of the protocol. Cells are sonicated to solubilize DNA fragments of appropriate length, incubated with an antibody targeting a protein of interest, immunoprecipitated, and washed. Crosslinking is reversed to yield chromatin enriched for sequences interacting with the protein of interest, and the DNA is prepared for sequencing with identifying indexing as necessary and finally sequenced. (B) Subway map for the semi-automated PerCell normalization analysis pipeline with cellular spike-in (red), experimental chromatin (blue), or the workflow with no spike-in option (yellow). With a streamlined user interface, our pipeline will automate the central processes for cellular spike-in chromatin normalization per cell, including downsampling for peak calling and generation of bigWig files. In our updated semi-automated pipeline, we include the option to either include or not include spike-in, which creates versatility and applicability for an increased range of researchers and projects.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep Methods