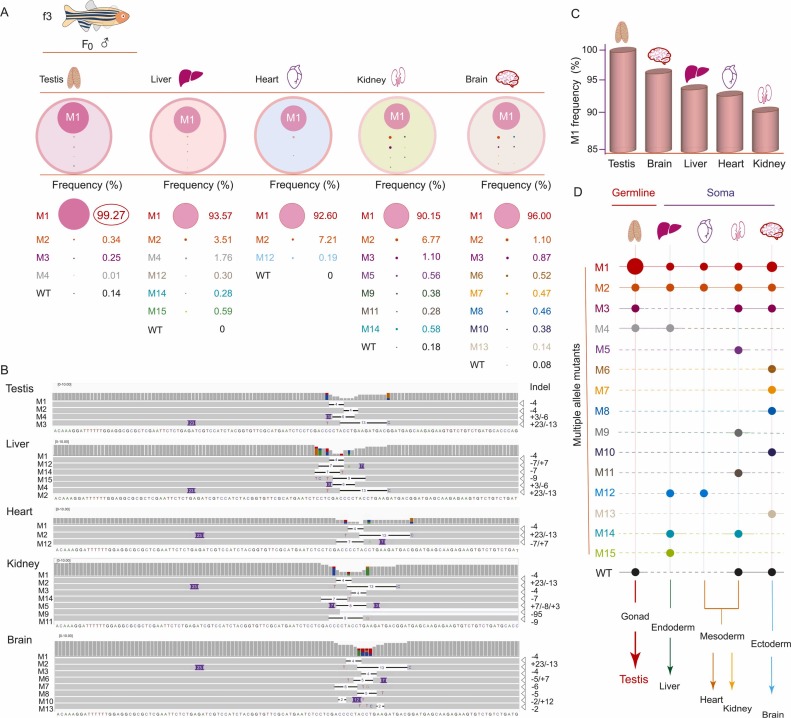
Fig. 4 Mapping of edited alleles among the 3 germ layers and future tissue types by deep sequencing. A. Allele typing by indels and frequency in testis and other somatic tissues (liver, heart, kidney, and brain) of the f3 founder. The cycle size in color represents the relative frequency of each allele. M(1,2,…15) indicates edited alleles. WT, wild type. B. Alignments of amplicon sequence variants by an integrative genomics viewer from deep sequencing of genomic DNA in the testis, liver, heart, kidney, and brain. Indels are shown in each edited sequence, and the base number in each indel is indicated on the right. The sequence of wild-type Sox9a is shown at the bottom of each alignment group. C. Percentage of frequency of allele M1 among tissues. D. Mapping of edited alleles and WT alleles among the 3 germ layers and future tissue types (testis and other somatic tissues). Dots in color indicate the alleles edited. WT, wild-type.
Reprinted from New biotechnology, , Yang, K., Cai, L., Zhao, Y., Cheng, H., Zhou, R., Optimization of genome editing by CRISPR ribonucleoprotein for high efficiency of germline transmission of Sox9 in zebrafish, , Copyright (2025) with permission from Elsevier. Full text @ N. Biotechnol.