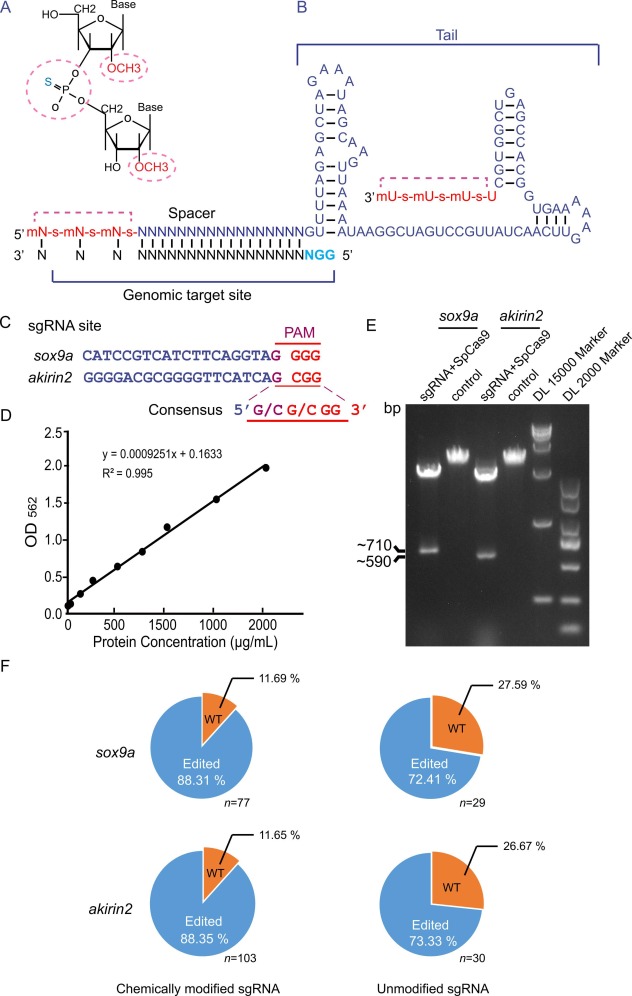
Fig. 1 The CRISPR RNP complex with chemically modified sgRNA cuts DNA in vitro and edits the zebrafish genome with high efficiency. A. Schematic diagram of chemical modification sites. The 2′-O-methyl and 3′ phosphorothioate modification sites are indicated in dotted cycles (red). B. The last 3 nucleotides at both ends of the RNA were chemically modified with 2′-O-methyl (m) and 3′ phosphorothioate (s). The last four bases are U at the 3′ end. Chemically modified nucleotides are highlighted in red. The sgRNA was designed with two hairpin structures in the 3′ tail. The target region is located at the 5′ end of the sgRNA. The pan site is denoted by the blue letter. C. Alignment of sgRNA sites highlights consensus sequence of PAM in sox9a and akirin2 genes. D. Quantification of the purified SpCas9 protein concentration determined by a Pierce BCA protein assay kit. E. RNP in vitro cutting efficiency. The target sequences (sox9a or akirin2) were used as the substrates for CRISPR RNP digestion. The cleavage products were examined by 1 % agarose gel electrophoresis. F. Statistical chart of genome editing proportion in zebrafish of the F0 generation. The number of F0 individuals is indicated in the sox9a and akirin2 groups. Unmodified sgRNA groups are controls for modified sgRNAs. WT, wild-type; Edited, gene-edited individuals.
Reprinted from New biotechnology, , Yang, K., Cai, L., Zhao, Y., Cheng, H., Zhou, R., Optimization of genome editing by CRISPR ribonucleoprotein for high efficiency of germline transmission of Sox9 in zebrafish, , Copyright (2025) with permission from Elsevier. Full text @ N. Biotechnol.