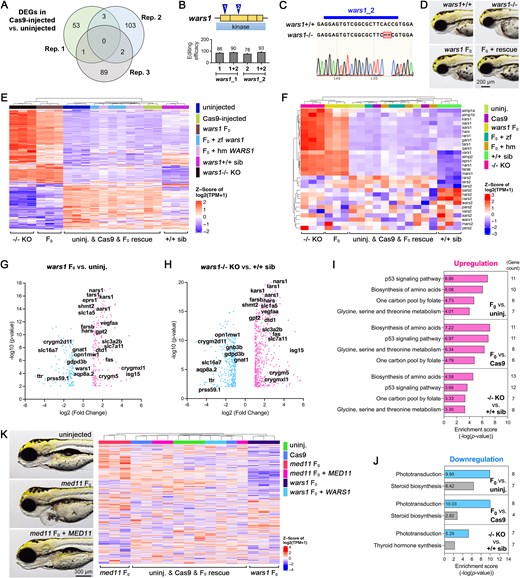
Fig. 5 Comprehensive transcriptomic analysis of uninjected and Cas9-injected control, F0, stable knockout, and rescued animals. (A) A Venn diagram compares DEGs in Cas9-injected versus uninjected embryos at 3 dpf. Three replicates indicate three independent runs of RNA-sequencing (RNA-seq), with each replicate containing biological triplicates (n = 6 embryos for each biological replicate). (B) Schematics of two gRNA target loci (blue triangle) and exons encoding kinase domain (yellow boxes) for the wars1 gene. The editing efficiency for each gRNA, injected either individually or in combination, is shown below. (C) Sequence alignment and chromatogram showing a 4bp deletion in the wars1 stable mutant generated by the wars1_2 gRNA, as revealed by Sanger sequencing. (D) Representative images of morphology for wars1 WT sibling (wars1+/+), homozygous knockout (wars1-/-), F0 and F0 rescued with human WARS1 mRNA embryos at 3dpf. Lateral view and anterior to the left. (E) A heatmap visualization of DEGs in uninjected (uninj.), Cas9 protein-injected (Cas9), wars1 F0, F0 rescued with zebrafish wars1 mRNA (F0 + zf), F0 rescued with human WARS1 mRNA (F0 + hm), wars1 WT sibling (+/+ sib) and homozygous knockout (-/- KO) animals from RNA-seq data. The genes used for heatmap visualization were a set of DEGs in the F0 versus uninjected group. (F) Another heatmap visualizes genes involved in aminoacyl-tRNA biosynthesis. (G and H) Volcano plots for wars1 F0 versus uninjected control and wars1-/- versus wars1+/+ sibling animals. Overlapped top DEGs are revealed on the figure. (I and J) KEGG pathway analysis shows DEGs enrichment in three groups (wars1 F0 versus uninjected, F0 versus Cas9, and wars1-/- versus wars1+/+ sibling) and is further divided into upregulation (I) and downregulation (J) groups. (K) Representative images of morphology for uninjected, med11 F0 and F0 rescued with human MED11 mRNA at 4 dpf are shown at the left panel. The heatmap at the right panel visualizes 1000 DEGs of uninjected, Cas9-injected, med11 F0, med11 F0 rescued with human MED11 mRNA, wars1 F0 and wars1 F0 rescued with human WARS1 mRNA animals at 3 dpf. Each group contains biological triplicates with six animals in each replicate. Note that one outlier in Cas9 group was removed in (E–J), and another in F0 + WARS1 group was removed in (K). For (A, E-K), the threshold cutoff was set to two-fold change (log2 fold change ≥ ±1) and a P-value less than 0.01.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nucleic Acids Res.