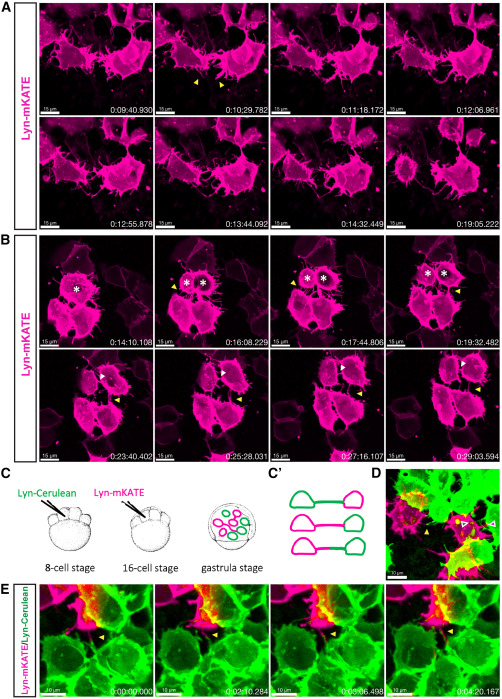
Fig. 2 Formation mechanisms of TNT-like connections (A) Time-lapse recording of the zebrafish embryo at 8 hpf shows two neighboring cells extending filopodia (yellow arrowheads) that subsequently merge into a single membranous connection that remains stable for at least 5 min. (B) Time-lapse recording of the zebrafish embryo at 8 hpf shows three neighboring cells forming TNT-like connections through cell dislodgement (yellow arrowhead) while the top cell is dividing (white asterisks). The daughter cells maintain connections with the lower cells for at least 15 min (yellow arrowhead), while a cytokinetic bridge between the daughter cells is also visible (white arrowhead). (C) Schematic representation of the strategy for labeling two cell populations: zebrafish embryo is injected with lyn-Cerulean mRNA into one of the central cells at the 8-cell stage, followed by injection at the 16-cell stage with lyn-mKATE mRNA, and then imaged at the gastrula stage (7–9 hpf). (C′)—Expected labeling outcomes: TNT-like structures formed by either one or both cell populations. (D) Representative image showing connections formed by either one (green and magenta arrowheads) or both (yellow arrowhead) cell populations. (E) Time-lapse recording of the zebrafish embryo at 8 hpf shows two cells belonging to different cell populations extending protrusions (yellow arrowhead) that elongate along each other. Scale bars are 15 μm for (A) and (B), 10 μm for (D) and (E), and time is indicated as h:mm:ss.
Reprinted from Developmental Cell, , Korenkova, O., Liu, S., Prlesi, I., Pepe, A., Albadri, S., Del Bene, F., Zurzolo, C., Tunneling nanotubes enable intercellular transfer in zebrafish embryos, , Copyright (2024) with permission from Elsevier. Full text @ Dev. Cell