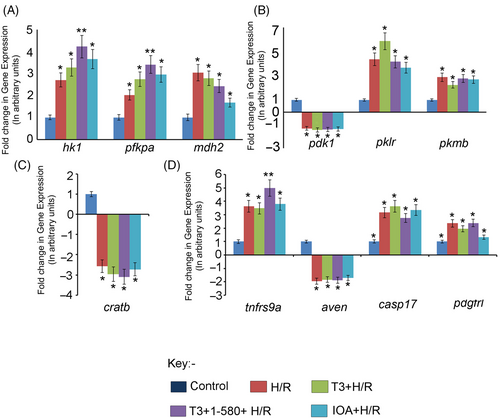
Fig. 10 Graph showing relative change in the expression of a few representative genes in different experimental groups as revealed by qRT-PCR analysis. (A) DEGs related to Glycolysis and Krebs Cycle—hexokinase 1 (hk1); phosphofructokinase, platelet a (pfkpa); malate dehydrogenase 2 and NAD (mitochondrial) (mdh2). (B) DEGs related to Pyruvate metabolism—pyruvate dehydrogenase kinase, isozyme 1 (pdk1); pyruvate kinase L/R (pklr) and pyruvate kinase M1/2b (pkmb). (C) DEG related to fatty acid oxidation—carnitine O-acetyltransferase b (cratb). (D) DEGs related to apoptosis and regeneration—tumour necrosis factor receptor superfamily, member 9a (tnfrsf9a); apoptosis, caspase activation inhibitor (aven); caspase 17, apoptosis-related cysteine peptidase (casp17) and platelet-derived growth factor receptor-like (pdgfrl). Data are shown as comparison between groups Con and H/R Group, Con and T3 + H/R Group, Con and T3 + 1–580 + H/R Group and Con and IOA+ H/R Group (*p < 0.05; **p < 0.01).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Wound Repair Regen.