|
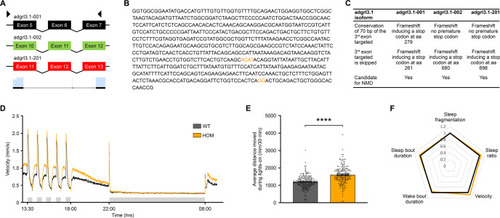
CRISPR-Cas9 mutagenesis and behavioral parameters of adgrl3.1−/− larvae. A Cartoon showing position of forward and reverse sgRNA constructs used to create the novel adgrl3.1−/− line. B The base pair sequence of adgrl3.1 that is deleted by sgRNA injection. The nucleotides shown in orange were not removed and so are retained in the mutant version of adgrl3.1. C The CRISPR guide RNAs were designed to target the different splice variants of adgrl3.1 found in the Ensembl database. (NMD: non-sense mediated decay). D Swim velocity of homozygous wild type (WT, n = 184) and homozygous adgrl3.1−/− (HOM, n = 179) larvae throughout the whole recording period. The data are pooled from four experiments. The time between 13:30 and 18:00 shows alternating intervals of 30-min lights-off (grey shaded bar) and lights-on phases, followed by constant light-on from 18:00 to 22:00. For the night time, from 22:00 to 08:00 the next morning, the lights were turned off (grey shaded bar). E The average distance moved for the five 30-min lights-off periods demonstrates that homozygous adgrl3.1−/− larvae moved significantly more than wild-type larvae. F Sleep parameters, expressed as fold change of wild-type larvae, were examined throughout the night, during 10-h lights-off phase. No significant differences between genotypes were observed in any of the five sleep parameters. * denote significant differences.
|