Fig. 2
- ID
- ZDB-FIG-221216-12
- Publication
- Spelat et al., 2022 - Metabolic reprogramming and membrane glycan remodeling as potential drivers of zebrafish heart regeneration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
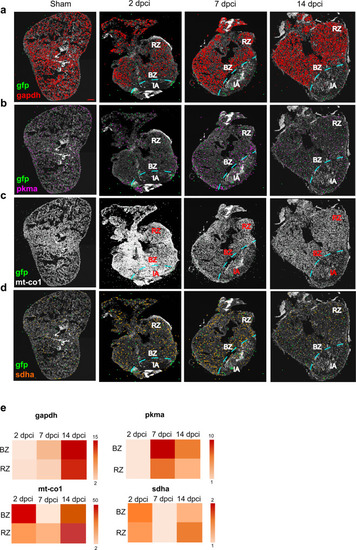
Cardiomyocyte targeted in situ sequencing.
A targeted multiplexed mRNA detection assay employing padlock probes, rolling circle amplification (RCA), and barcode sequencing was applied to detect the spatial distribution of a gapdh b pkma c mt-co1 and d sdha in sham and at 2, 7 and 14 dpci. All the experiments were performed on cmlc2 zebrafish transgenic strain, where a green fluorescent protein (gfp) gene was inserted under the promoter of cardiomyocyte-specific gene cardiac myosin light chain 2. For this reason, gfp was chosen as a cardiomyocyte marker. IA injured area, BZ border zone, RZ remote zone. Scale bar = 100 μm. e Heatmaps showing the expression comparison of gapdh, pkma, mt-co1 and sdha between BZ and RZ at each time point. Heat legend represents the relative quantification of the spot number and it is represented as fold change. Two biological replicates were analyzed. |