- Title
-
Dimensionality reduction of calcium-imaged neuronal population activity
- Authors
- Koh, T.H., Bishop, W.E., Kawashima, T., Jeon, B.B., Srinivasan, R., Mu, Y., Wei, Z., Kuhlman, S.J., Ahrens, M.B., Chase, S.M., Yu, B.M.
- Source
- Full text @ Nat Comput Sci
|
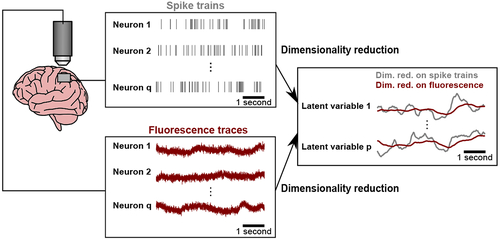
A key property of calcium imaging is the slow decay of the measured fluorescence (left panel, maroon) after each spiking event (left panel, grey). If ignored, the calcium decay could introduce temporal correlations in the estimated latent variables (right panel, maroon), where those temporal correlations would not be present had we estimated the latent variables from the underlying spike trains (right panel, grey). |
|
|
|
|
|
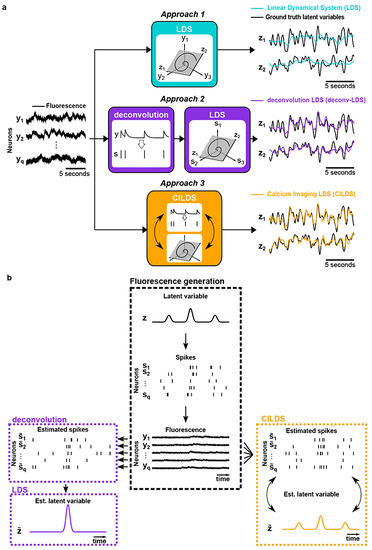
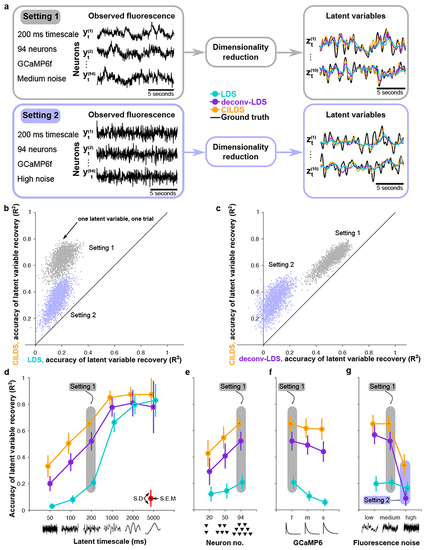
Here we compare the performance of three methods at recovering ground truth latent variables in simulation: one with deconvolution and latent dynamics (CILDS), one with de-convolution but no latent dynamics (CIFA), and one with no deconvolution and no latent dynamics (FA). The simulation parameters are GCaMP6f with 94 neurons and medium noise, as in |
|
|
|
|