PUBLICATION
EmbryoMiner: A new framework for interactive knowledge discovery in large-scale cell tracking data of developing embryos
- Authors
- Schott, B., Traub, M., Schlagenhauf, C., Takamiya, M., Antritter, T., Bartschat, A., Löffler, K., Blessing, D., Otte, J.C., Kobitski, A.Y., Nienhaus, G.U., Strähle, U., Mikut, R., Stegmaier, J.
- ID
- ZDB-PUB-180421-10
- Date
- 2018
- Source
- PLoS Computational Biology 14: e1006128 (Journal)
- Registered Authors
- Mikut, Ralf, Otte, Jens, Strähle, Uwe, Takamiya, Masanari
- Keywords
- none
- MeSH Terms
-
- Cell Movement
- Software
- Embryonic Stem Cells/cytology
- Embryonic Development
- Animals, Genetically Modified
- PubMed
- 29672531 Full text @ PLoS Comput. Biol.
Abstract
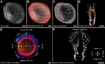
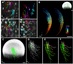
State-of-the-art light-sheet and confocal microscopes allow recording of entire embryos in 3D and over time (3D+t) for many hours. Fluorescently labeled structures can be segmented and tracked automatically in these terabyte-scale 3D+t images, resulting in thousands of cell migration trajectories that provide detailed insights to large-scale tissue reorganization at the cellular level. Here we present EmbryoMiner, a new interactive open-source framework suitable for in-depth analyses and comparisons of entire embryos, including an extensive set of trajectory features. Starting at the whole-embryo level, the framework can be used to iteratively focus on a region of interest within the embryo, to investigate and test specific trajectory-based hypotheses and to extract quantitative features from the isolated trajectories. Thus, the new framework provides a valuable new way to quantitatively compare corresponding anatomical regions in different embryos that were manually selected based on biological prior knowledge. As a proof of concept, we analyzed 3D+t light-sheet microscopy images of zebrafish embryos, showcasing potential user applications that can be performed using the new framework.
Genes / Markers
Expression
Phenotype
Mutations / Transgenics
Human Disease / Model
Sequence Targeting Reagents
Fish
Orthology
Engineered Foreign Genes
Mapping