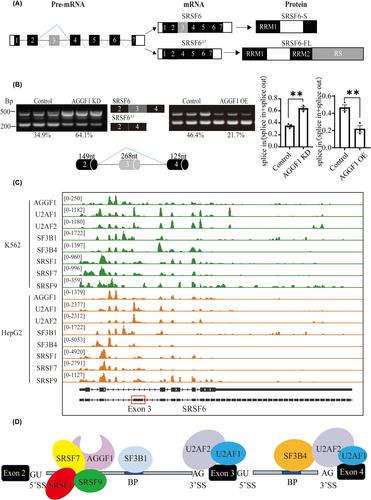
Fig. 4 AGGF1 promotes exon 3 skipping of SRSF6 during alternative splicing. (A) Structure of SRSF6 pre-mRNA, two mature spliced RNA transcripts (SRSF6 with all exons and SRSF6Δ3 with deletion of exon 3), and two protein isoforms. Transcript SRSF6 is translated into a shorter SRSF6 protein with RS and part of RRM2 domain deleted, whereas transcript SRSF6Δ3 is translated into the full-length SRSF6 protein. (B) Semi-quantitative RT-PCR analysis to confirm the AS event of exon 3 skipping of SRSF6. Two alternatively spliced SRSF6 RNA transcripts are shown as two distinct bands on agarose gels, and were both abundant in HPAEC (control). AGGF1 knockdown or overexpression of FLAG-tagged AGGF1 altered AS event of exon 3 skipping of SRSF6 (n = 3 samples/group; **p < .01 vs. Control). (C) Genome browser view of the SRSF6 locus showing eCLIP signal for AGGF1, U2AF2, U2AF1, SF3B1, SF3B4, SRSF1, SRAF7, SRSF9 in K526 and HepG2 cells. eCLIP data were downloaded from ENCODE database (https://www.encodeproject.org/). (D) Results of eCLIP data from (C) showing the binding sites for AGGF1 and other splicing regulatory factors during spliceosome complex assembly at the 5′-splice site (5′SS), 3′-splice site (3′SS), branch point site (BP) in the pre-mRNA region spanning exons 2 to 4.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ FASEB J.